Abstract
To capture global responses to metal poisoning and mechanistic insights into metal toxicity, gene expression changes were evaluated in whole adult male zebrafish following acute 24 h high dose exposure to three metals with known human health risks. Male adult zebrafish were exposed to nickel chloride, cobalt chloride or sodium dichromate at concentrations corresponding to their respective 96 h LC20, LC40 and LC60 (i.e. 96 h concentrations at which 20%, 40% and 60% lethality is expected, respectively). Histopathology was performed on a subset of metal-exposed zebrafish to phenotypically anchor transcriptional changes associated with each metal exposure. Here we describe in detail the contents and quality controls for the gene expression and other data associated with the study published by Hussainzada and colleagues in BMC Pharmacology and Toxicology (Hussainzada et al., 2014) with the data uploaded to Gene Expression Omnibus (accession number GSE50648).
Keywords: Zebrafish, Whole organism, Nickel, Chromium, Cobalt, Toxicogenomics
| Specifications | |
|---|---|
| Organism/cell line/tissue | Danio rerio (Tübingen strain) |
| Sex | Male |
| Sequencer or array type | Agilent 4 × 44 K custom design (see Microarray hybridization for details) |
| Data format | Raw data: .gpr files; normalized data: SOFT, MINML, and TXT |
| Experimental factors | Metal exposure vs. unexposed normal |
| Experimental features | Male adult zebrafish were exposed to nickel chloride, cobalt chloride or sodium dichromate concentrations corresponding to their respective 96 h LC20, LC40 and LC60. Histopathology was performed on a subset of metal-exposed zebrafish to phenotypically anchor transcriptional changes associated with each metal as determined by microarray analysis. |
| Consent | None |
| Sample source location | In-house aquaculture at US Army Center for Environmental Health Research, Fort Detrick, Frederick, MD, USA |
Direct link to deposited data
Deposited data can be found here: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50648.
Experimental design, materials and methods
Experimental overview
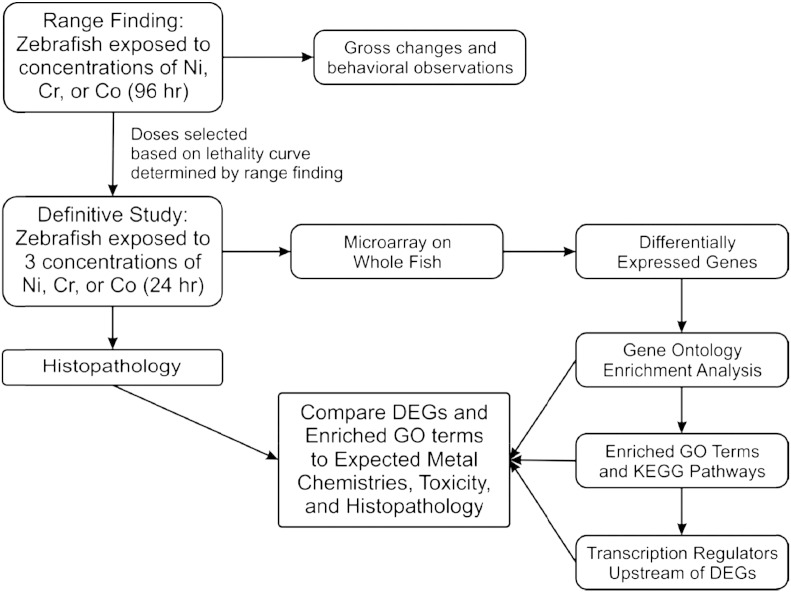
Fig. 1 shows the design scheme.
Fig. 1.
Fish exposures
Briefly, adult, male zebrafish were exposed to concentrations of each metal necessary for 20% (LC20; low), 40% (LC40; mid) and 60% (LC60; high) lethality based on prior 96 h range-finding experiments. Exposures were conducted for 24 h using control (no toxicant) plus the high, mid- and low concentrations of each metal (Table 1). The intent was to evaluate levels of toxicant sufficient to induce a measurable response without producing lethality at 24 h. All metal concentrations in the test tanks were verified both before and after exposures. Initially 25 adult (6–9 months) presumptive male zebrafish were selected per condition to ensure that 20 male fish were available for subsequent microarray analysis and histopathology. At termination of each study, five fish per condition were sacrificed by terminal dose of MS222 and then immediately preserved in a modified Davidson's solution for histological examination. For transcriptional analysis, the remaining 20 zebrafish were sacrificed by terminal dose of MS222 and then immediately immersed whole in liquid nitrogen and stored at − 80 °C until RNA processing.
Table 1.
Nominal and actual concentrations (mg/L) of metals used in 24-hour zebrafish exposures.
| Treatment | Nominal LC20 (low) | Measured LC20 (low) | Nominal LC40 (mid) | Measured LC40 (mid) | Nominal LC60 (high) | Measured LC60 (high) |
|---|---|---|---|---|---|---|
| NiCl2 | 45 | 42.4 | 54 | 51.0 | 62 | 64.0 |
| CoCl3 | 39 | 39.7 | 50 | 46.3 | 65 | 59.5 |
| Na2Cr2O7 | 53 | 56.5 | 65 | 69.9 | 76 | 80.6 |
From Hussainzada et al. (2014).
RNA processing
Flash frozen whole fish were individually pulverized under liquid nitrogen using a SPEX 6750 freezer mill (SPEX Sample Prep, Metuchen, NJ). Six milliliters of Trizol® (Invitrogen, Carlsbad, CA) was added to the pulverized material, vortexed well and transferred to a 10 mL Dounce homogenizer. An additional 2 mL of Trizol® was added to remove any remaining pulverized material from the initial tube and transferred to the same Dounce. The pulverized material was further homogenized by hand with 10 strokes using Pestle A followed by 10 strokes using Pestle B. After homogenization, the manufacturer's suggested protocol was followed for RNA extraction with an extra clarification centrifugation step to remove bone, scales, and other insoluble debris from the homogenate. Total RNA isolated via Trizol® was then subjected to further purification using RNeasy® Midi kits (Qiagen, GmbH, Germany) to remove any residual salts or organic solvents. Quality and quantity of isolated total RNA were analyzed using an Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA) and verified with the NanoDrop ND-1000 Spectrophotometer (NanoDrop, Wilmington, DE). Total RNA showing signs of degradation or a RIN value < 8 was not included in further analysis.
Screening
Prior to performing microarray analysis, the RNA was screened against a primer panel to verify that the RNA was isolated from male fish only. An aliquot of each total RNA sample was transcribed to cDNA using the Advantage® RT-for-PCR Kit (Clontech, Mountain View, CA). Amplification and detection were performed on an MJ Opticon® 2 (BioRad, Hercules, CA) using SYBR® Green PCR Master Mix (Applied Biosystems, Carlsbad, CA) with primers designed to detect transcripts coding for vitellogenin 1 (vit1, expressed only in female liver) and glyceraldehyde 3-phosphate dehydrogenase (gapdh). gapdh was used as an internal control for normalizing sample concentrations. Any sample which showed the presence of female RNA (i.e. high levels of vit1) was eliminated from further analysis.
Microarray hybridization
In order to maximize statistical power while minimizing cost, equal amounts of total RNA from four or five individual fish from each exposure condition in each of the four exposure experiments were combined to create four biological replicate pools. Each biological replicate pool was hybridized to a separate microarray for a total of 16 microarrays (i.e. four control replicates, four low dose replicates, four mid-dose replicates, and four high dose replicates).
For this study, custom designed microarrays were used. The arrays were designed in-house using the eArray microarray design tool (https://earray.chem.agilent.com/earray/; Agilent Technologies, Inc.) using zebrafish gene targets derived from Ensembl build 46 (Zv7 genome build) and Vega build 26. Each array contains 44,000 60-mer oligonucleotides representing 21,904 zebrafish gene targets with two probes per transcript wherever possible; only 94 transcripts had a single probe. Probes were designed using only genes which are annotated in published databases. All arrays were manufactured by Agilent.
All microarrays were processed following Agilent's One-Color Microarray-Based Gene Expression Analysis Protocol (Version 5.5, February 2007) for processing 4 × 44 K array slides. An initial 1 μg pooled total RNA input was used with an 18 hour overnight hybridization at 65 °C. A final wash using the Stabilization and Drying solution (Agilent Technologies, Inc.) was included after the required specificity washes in order to prevent any ozone related degradation of signal prior to scanning the arrays. Arrays were scanned on a GenePix Autoloader 4200 AL (Molecular Devices, Union City, CA) using the 532 nm (green) laser at 45% with the green filter, PMT = 400, and the scan resolution = 5 μm. Raw images were processed using GenePix Pro 6.0 (Molecular Devices) with the following settings: GAL file = 0177662_D_20070905_update8.gal; local background subtraction; background subtraction: width of background = 2 feature diameters; resize features: minimum diameter = 80%; resize features: maximum diameter = 110%; feature movement: maximum translation = 20 μm; and CPI threshold = 0. These setting were determined to be optimal for grid alignment based on optimization experiments performed in house. Prior to performing final quality control checks, arrays were visually inspected to verify grid alignment and the absence of any defects. All microarray data from this study have been deposited in NCBI's Gene Expression Omnibus [1] under the accession number GSE50648.
Quality control & normalization
GenePix Pro offers intensity data in several alternate forms. For this study [2], median signal intensity and signal-to-noise ratio (SNR) were used. Median signal intensity is calculated as the median value of all the pixels that are completely with the feature indicator; any pixel touching the boundary is excluded. This software calculates SNR as the difference between median spot intensity signal and median background divided by the standard deviation of the background signal.
Median signal intensity array data was imported into Partek Genomics Suite (version 6.6; Partek Inc., St. Louis, MO) for additional analysis. To verify performance of the replicates, Pearson's R2 was calculated for each set of replicates within each toxicant group. Any arrays with R2 < 0.95 were excluded from analysis. All arrays included in the final analysis had R2 > 0.97 with their replicates.
To further increase confidence, only unsaturated probes (i.e. probes with signal intensity less than 65,535) with a SNR greater than or equal to three (SNR ≥ 3) were selected for analysis. The SNR data was imported into Partek Genomics Suite; a transposed spreadsheet was created and then transferred to Microsoft Excel (Microsoft Corporation, Redmond, WA) where a list was created of all probe sets with a SNR ≥ 3 in all replicates of at least one condition. This list was then used to filter the median signal intensity data in Partek. Only unsaturated probes with SNR ≥ 3 were selected for analysis which yielded a subset of 15,818 probes which map to 7909 genes. Quantile normalization across arrays was performed to control for any inter-array variability then these normalized probe intensities were log transformed.
Basic analysis
Using Partek Genomics Suite, three sets of ANOVAs (one set for each metal) were performed to identify probes that were differentially expressed between each metal exposure group and its control group. The ANOVAs included terms for treatment (exposed or unexposed) and concentration (control, low, mid, or high) and an interaction term for “treatment ∗ concentration”. For each chemical, contrasts were performed to determine significance between each dose group and its respective control. A step-up Benjamini and Hochberg false discovery rate (FDR) of 0.01 was used to select differentially expressed probes. An FDR α = 0.01 was selected as a cut-off for the combined nickel, chromium, and cadmium datasets including all replicate pools. Probes not meeting the FDR α = 0.01 threshold were eliminated from further analysis. The resulting list was further refined by submitting it to a second filter which specified a 1.8-fold change between exposed versus control. Only transcripts with probes which passed both filters were included in the final analysis. For all transcripts with two probes, fold changes for each probe pair were averaged to generate a single value for each transcript. For all transcripts with a single probe, fold change data stood as is.
Exposure to chromium, cobalt, or nickel significantly altered the expression of 696, 461, and 287 genes respectively (Fig. 2).
Fig. 2.
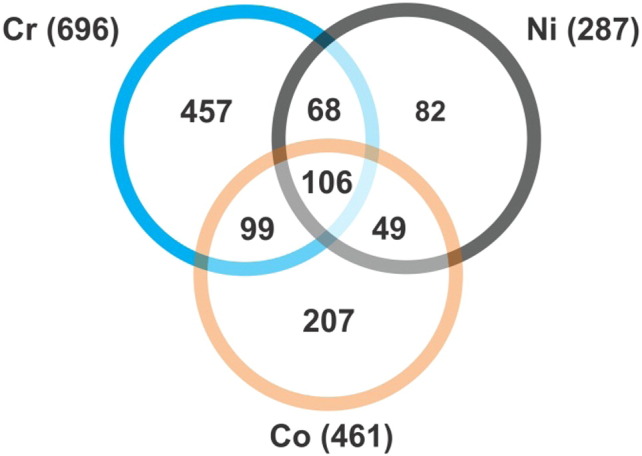
Total number of genes significantly (FDR = 0.01) altered by at least 1.8 fold in response to chromium (Cr), cobalt (Co), or nickel (Ni) versus untreated controls.
From Hussainzada et al. (2014).
Discussion
We describe here a unique dataset of male zebrafish transcriptomic responses to acute metal toxicity. This dataset is composed of genome-wide gene expression data measured using a custom Agilent array platform. The gene expression data is of high quality and consistent with histopathologic data collected during this study [2]. Using this data, we identify changes in gene expression of groups of genes consistent with adaptive responses induced by exposure to nickel, cobalt, or chromium including acute phase response, cell cycle regulation, apoptosis, and metabolic depression.
Disclaimer
Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army. Research was conducted in compliance with the Animal Welfare Act, and other Federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals (NRC 2011) in facilities that are fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care, International. The research described herein was sponsored by the U.S. Army Medical Research and Materiel Command, Military Operational Medicine Research Program. Citations of commercial organizations or trade names in this report do not constitute an official Department of the Army endorsement or approval of the products or services of these organizations.
This project was supported in part by an appointment to the Research Participation Program for the U.S. Army Medical Research and Materiel Command administered by the Oak Ridge Institute for Science and Education through an agreement between the U.S. Department of Energy and U.S. Army Medical Research and Materiel Command.
References
- 1.Barrett T., Suzek T.O., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R. NCBI GEO: mining millions of expression profiles — database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hussainzada N., Lewis J.A., Baer C.E., Ippolito D.L., Jackson D.A., Stallings J.D. Whole adult organism transcriptional profiling of acute metal exposure in male zebrafish. BMC Pharmacol. Toxicol. 2014;15(1):15. doi: 10.1186/2050-6511-15-15. [DOI] [PMC free article] [PubMed] [Google Scholar]