Abstract
The protein kinase C (PKC) activator phorbol 12-myristate 13-acetate (PMA) induces transition of the epithelial MCF-7 cell line to a mesenchymal phenotype. A subset of the resulting mesenchymal cells has surface markers characteristics of a cancer stem cell (CSC) population. We profiled the transcriptome changes associated with the epithelial to mesenchymal transition and those that occurred in the CSC subset. Using a siRNA knockdown strategy, we examined the extent to which these changes were dependent on the PKC family member, PKC-θ. The importance of the cytoplasmic signaling role of this kinase is well established and in this study, we have shown by PKC-θ ChIP-sequencing analysis that this kinase has a dual role with the ability to also associate with chromatin on a subset of PKC-θ dependent genes. In the associated manuscript (Zafar et al., 2014 [5]) we presented evidence for the first time showing that this nuclear role of PKC-θ is also important for gene induction and mesenchymal/CSC phenotype. Here we describe the analysis associated with the transcriptome and ChIP-seq data presented in Zafar et al. (2014) [5] and uploaded to NCBI Gene Expression Omnibus (GSE53335).
Keywords: ChIP-seq, Microarrays, Stem cell
Specifications
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens/MCF-7 cell line/mammary adenocarcinoma |
| Sex | Female |
| Sequencer or array type | Microarray: Affymetirx Human Gene 1.0 ST and Human Gene 2.0 ST |
| ChIP-seq: Illumina Hi-Seq 2000 | |
| Data format | Microarray raw data: CEL files |
| ChIP-seq raw data: FASTQ files | |
| ChIP-seq processed data: wig files and bed files | |
| Experimental factors | PMA treatment and siRNA transfection |
| Experimental features | Microarray gene expression profiling to identify genes that are dependent on PKC-theta (PRKCQ). |
| ChIP-seq to map genomic sites that are bound by PKC-theta | |
| Consent | N/A |
| Sample source location | N/A |
Direct link to deposited data
Experimental design, materials and methods
Cell culture and RNA isolation
The MCF-7 cell line was cultured at 37 °C in low glucose DMEM (Gibco) supplemented with fetal calf serum (10%), l-glutamine (2 mM), and penicillin and streptomycin (0.1%). Non-stimulated (NS) MCF-7 cells were stimulated for 60 h with phorbol 12-myristate 13-acetate (PMA, 0.65 ng/ml, Sigma-Aldrich) to generate the whole stimulated population (WP) before FACS sorting using antibodies against CD44-APC (559942, BD Biosciences) and CD24-PE (555428, BD Biosciences) to give the cancer stem cell (CSC) and non-cancer stem cell (NCSC) populations. Hoechst 33258 (561908, BD Biosciences) was used while sorting to exclude dead cells. The CSC population was gated as CD44 high and CD24 low, while the NCSC population was the remaining stimulated cells. Total RNA was isolated from these cells using TRIzol (Invitrogen), labeled and hybridized to Affymetrix Hugene 1.0 ST (at the Ramaciotti Centre for Genomics, NSW).
MCF-7 cells were transfected with PKC-θ siRNA (sc-36252) or mock siRNA (Fluorescein conjugate, sc-36869) (20 nM, Santa Cruz Biotechnologies) using Lipofectamine 2000 (Invitrogen). Transfected cells were left for 48 h before being stimulated with PMA. This resulted in four samples; mock non-stimulated (mock NS), mock stimulated (mock ST), PKC siRNA non-stimulated (pkc NS), and PKC siRNA stimulated (pkc ST). Total RNA was isolated from these cells using TRIzol, labeled and hybridized to Affymetrix Hugene 2.0 ST.
Microarray analysis
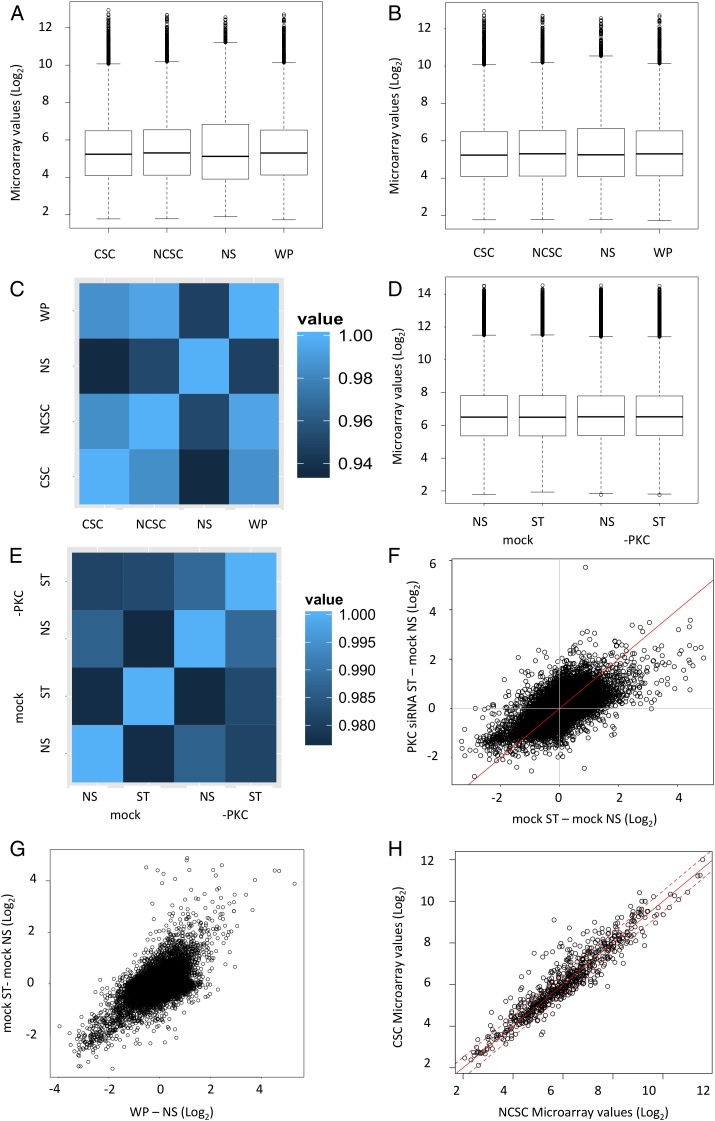
When Robust Multiarray average (RMA) normalization (Affymetrix Powertools) was used on the Hugene 1.0 ST arrays, the NS sample values displayed a different distribution to the WP, CSC and NCSC samples (Fig. 1A). Loess normalization (using the LPE package in R) was used to ‘correct’ the NS distribution (Fig. 1B). The NS sample still showed the least similarity (Fig. 1C) to the others but this was expected as the other samples were all from stimulated cells. As the WP population consists of 89.5% of NCSC cells and 10.5% CSC cells, its slightly higher correlation with the NCSC sample was as expected (Fig. 1C).
Fig. 1.
Value distributions and correlations of the microarray data. Box and whisker plots show the RMA normalized distributions of the Log2 values before (A) and after (B) loess normalization for the microarrays on nonstimulated (NS), PMA stimulated (WP) and PMA stimulated cells sorted into cancer stem cell (CSC) and non-cancer stem cell (NCSC) populations. RMA normalization was used for the microarrays performed on RNA from cells treated with mock or PKC-θ siRNA (-PKC) and stimulated with PMA (ST) (D). The microarray samples were compared to each other using Pearson correlations (C, E). Using values from the microarray the changes in expression between PKC-θ siRNA ST cells and mock NS cells were compared to those between mock ST cells and mock NS cells (F) to determine if any changes were PKC-θ dependent. Changes in expression between mock ST and mock NS cells from the Hugene 2.0 ST array were compared to those from the WP and NS cells from the Hugene 1.0 ST array (G). Differences in expression between the CSC and NCSC cell populations were compared for the PKC-θ sensitive genes (H) to determine if any were expressed more highly in the CSC population. The unbroken red line indicates x = y, while broken red lines indicate Log2 0.5 differences.
Hugene 2.0 arrays were simply RMA normalized (Affymetrix Powertools) (Fig. 1D). The two mock samples had lower correlation scores than the PKC-θ siRNA samples, indicating that PKC theta plays an essential role in at least a proportion of the gene expression changes that occurred with stimulation (Fig. 1E). A comparison of the changes measured in the mock ST samples compared to the in the PKC-θ siRNA ST sample, against the mock NS sample revealed that while PKC-θ was necessary for the increase and decrease of many genes it's inhibition did not abrogate all changes (Fig. 1F).
Genes were judged to be changed in expression if their Log2 difference was at least 0.5. 1356 genes were identified as PKC-θ sensitive in that they were induced by stimulation in the mock siRNA sample but were expressed less in the stimulated PKC-θ siRNA sample than the stimulated mock siRNA sample. This induction by PMA and sensitivity to PKC-θ was confirmed for 10 (of 10 tested) transcripts by quantitative real-time PCR [5].
To determine which of the PKC- sensitive genes were expressed more in CSC we had to compare probesets on the Hugene 1.0 ST and 2.0 ST arrays. The probesets were matched largely by gene symbol but were also considered to represent the same transcript if the chromosome, transcription start site and strand were identical. Using this approach only 42% of the probesets on the Hugene 2.0 ST array had comparable probesets on the Hugene 1.0 ST array. First we compared the two stimulations, the mock NS and mock ST comparison performed on the Hugene 2.0 arrays and the NS and WP comparison performed on the Hugene 1.0 ST arrays (Fig. 1G). Of the probesets comparable on both arrays, 672 were up-regulated with stimulation in both assays and this was 49.6% of those from the mock ST to mock NS comparison. When the 2 sets of Log2 differences were compared the Pearson correlation coefficient was 0.583. Whether differences were due to biological variation, the different probes used or the mock transfection was unclear. Of those represented on the Hugene 1.0 ST array, 134 (17.8%) of PKC-θ sensitive gene cohort were enriched in CSC compared to NCSC (Fig. 1H). Thus together the two microrarray studies allowed us to identify genes associated with PKC-θ induced CSC formation.
Chromatin immunoprecipitation-sequencing (ChIP-seq)
Non-stimulated (NS) and PMA stimulated (ST) MCF-7 cells were fixed with paraformaldehyde (1%) for 10 min at room temperature and lysed with the Upstate ChIP kit and sonication according to the manufacturer's instructions. Lysates were immunoprecipitated with anti-PKC-θ (5 μg, sc-212, Santa Cruz) and Protein A magnetic beads (Millipore).
The resulting ChIP-DNA (10 ng) was used with the NEBNext ChIP-Seq Library Prep for Illumina (New England BioLabs Inc, NEB#E6240L) according to the manufacturer's instructions. The kit consists of an End Repair Enzyme kit, Klenow fragment for dA-tailing, Quick T4 DNA ligase (with NEBNext adaptor primers) and NEBNext High-Fidelity 2× PCR Master mix, Universal PCR primer (25 μM), and index primer (25 μM, New England BioLabs Inc, NEBNext Multiplex Oligos for Illumina, NEB#E7335L) for multiplexing. A total of 15 cycles were used for PCR amplification of the adaptor-ligated DNA. Fragments were size selected to be between 175 and 225 bp using AMPure XP beads (Beckman Coulter, Inc., Part #: A63881). The quality of the ChIP-DNA library was assessed on a Bioanalyzer (Agilent). A total of 2 nM was captured on the Illumina flow cell for cluster generation and sequenced on the Illumina HiSeq2000 using a 50 bp run at the Ramaciotti Centre for Genomics.
Bioinformatics for ChIP sequencing
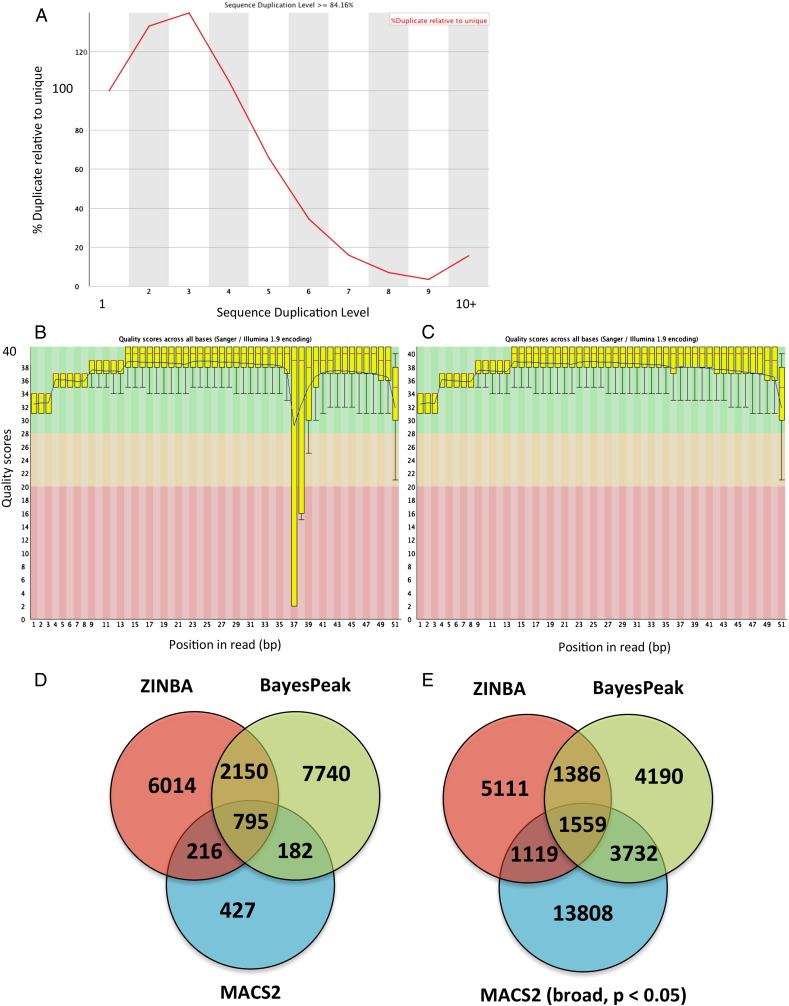
Total input (TI) samples were sequenced for both the NS and ST samples. Each of the PKC-θ and TI samples were run on 2 lanes. The quality of the reads was first checked with FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). All quality controls were passed except for high sequence duplication levels in the PKC-θ ST sample (Fig. 2A) and a problem at base 37 was noted for one lane (Fig. 2B) but not the other (Fig. 2C). The adapter sequences were trimmed using Cutadapt (http://code.google.com/p/cutadapt/) then mapped to the human genome (Hg19) using local alignment in Bowtie 2 (–local -D 20 -R 3 -N 0 -L 20 -i S,1,0.50) [3]. Only reads that uniquely mapped to the genome (46% for the PKC-θ ST samples) were used for subsequent analysis (obtained using awk ‘$13!~/XS/’). At this point data from the 2 lanes were combined and duplicate reads were removed using Picard (http://picard.sourceforge.net, default parameters). As expected from the FASTQC report the PKC-θ ST sample had a high number of duplicate reads (81%), compared to the PKC-θ NS (40%) and TI ST (28.7%). The resulting reads totaled 14,108,692 and 4,645,493 from the NS and ST MCF7 PKC-θ ChIP samples and 23,370,927 and 21,886,666 reads from the respective TI samples.
Fig. 2.
Quality control of sequencing and comparison of peak calling programs. Sequencing of libraries from PKC-θ bound DNA in MCF-7 cells stimulated with PMA, revealed high levels of sequence duplication (A) that had to be removed using Picard. One of the replicate lanes showed poor quality scores for the 37th base pair of reads (B) but the other replicate lane did not (C). Different numbers of PKC-θ enriched regions were called against total input for the stimulated cells, when using three programs; ZINBA (p value < 0.05), BayesPeak (posterior probability > 0.999) and MACS2 (q value < 0.01, D or q value < 0.05 and -broad, E).
PKC-θ enriched regions were called for the PKC-θ ST sample against the TI ST sample using a p-value cutoff of 0.05 for the Zero Inflated Negative Binomial Algorithm (ZINBA) ([4], extension = 200, refine peaks = 1, broad = F), a posterior probability cutoff of 0.999 for BayesPeak ([2], method = “lowerbound”) and MACS2 [6]. For MACS2 the parameters used were ‘(–keep-dup all -g hs -s 50 –bw 200 -q 0.05)’ but the resulting peaks were then filtered for q value < 0.01. Filtering on the q value after MACS2 had been used resulted in more peaks than if − q was set to 0.01. For representation in UCSC, reads were extended by 200 bp and HTSeq [1] was used to create wig files. Of the 2945 regions called as enriched by both BayesPeak and ZINBA, 795 (27%) were also found by MACS2 (Fig. 2D). However MACS2 did not call a PKC-θ enriched region at the BHLHE40 promoter, which was found to give the strongest enrichment in ChIP-PCR. Using the broad option for MACS2, and setting the p value cutoff to < 0.05 resulted in 20,218 peaks being called and a greater overlap with the other programs, especially BayesPeak (Fig. 2E). However, the broad option still did not call the BHLHE40 promoter peak.
To compare the NS and ST samples, only the peaks called by both BayesPeak and ZINBA from the ST sample were used and the number of reads for each of these peaks in the two samples was counted in R. The number of reads per peak was normalized to the reads per million (uniquely) mapped reads. As noted in [5] the resulting regression result was ST = 1.241 ∗ NS + 0.9575. The counts were not normalized further as ChIP-PCR for 6 (of 8 tested) of these regions confirmed a statistically significant increase in PKC-θ binding in ST compared to NS, and biologically it was not expected that the samples would have similar PKC binding levels [5].
As reported in [5] overlay of the transcriptome and ChIP-seq data identified 62 direct PKC-θ targeted genes whose induction with PMA was also dependent on PKC-θ. Importantly, many of these genes are known key genes previously implicated in epithelial to mesenchymal transition and formation of CSC [5].
Acknowledgments
We would like to thank Harpreet Vohra and Michael Devoy for the support with the FACS sorting and the financial support of the Endeavour Postgraduate Award (number 641_2008) and The Australian National University of Canberra PDF fellowship scheme.
References
- 1.Anders S., Pyl P.T., Huber W. HTSeq — a python framework to work with high-throughput sequencing data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cairns J., Spyrou C., Stark R., Smith M.L., Lynch A.G., Tavare S. BayesPeak — an R package for analysing ChIP-seq data. Bioinformatics. 2011;27:713–714. doi: 10.1093/bioinformatics/btq685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rashid N.U., Giresi P.G., Ibrahim J.G., Sun W., Lieb J.D. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 2011;12:R67. doi: 10.1186/gb-2011-12-7-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zafar A., Wu F., Hardy K., Li J., Tu W.J., McCuaig R., Harris J., Khanna K.K., Attema J., Gregory P.A., Goodall G.J., Harrington K., Dahlstrom J.E., Boulding T., Madden R., Tan A., Milburn P.J., Rao S. Chromatinized PKC- directly regulates inducible genes in epithelial to mensenchymal transition and breast cancer stem cells. Mol. Cell. Biol. 2014;34:2961–2980. doi: 10.1128/MCB.01693-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]