Abstract
Appropriate gene expression patterns form the basis for bone microvascular endothelial cells' function in femoral head. Although previous studies have elucidated the impact of glucocorticoids on these cells' specific gene expression the exact differential transcriptomes and comprehensive gene expression profiles remain unknown. Using microarray-based platforms we investigated the transcriptome patterns before and after hydrocortisone administration of bone microvascular endothelial cells from human femoral head. Our results highlight the involvement of development differentiation and apoptosis in the bone microvascular endothelial cells. Elucidation of differential gene expression before and after hydrocortisone administration emphasizes the importance of regulatory networks to gene co-expression within biological processes induced by glucocorticoids. With Benjamini–Hochberg characterization we identified 73 up-regulated and 166 down-regulated long noncoding RNAs the expression of 107 of which significantly correlated with 172 mRNAs after administration of hydrocortisone. Transcriptome analysis of bone microvascular endothelial cells from human femoral head samples is highly informative because it is deduced from data comprised of large number of genes expressed above background. The data have been submitted to the repository of Gene Expression Omnibus (Series GSE60332).
Keywords: Glucocorticoids-induced lesion, Bone microvascular endothelial cells, Long noncoding RNAs, Messenger RNAs, Microarray
| Specifications | |
|---|---|
| Organism/cell line/tissue | Bone microvascular endothelial cells from femur head of Homo sapiens |
| Sex | One male and seven females |
| Sequencer or array type | GPL19072 Agilent-052909 CBC_lncRNAmRNA_V3 (Probe name version) |
| Data format | Raw data |
| Experimental factors | Treated vs. untreated. Cells are duplicated to 60 mm gelatin-coated culture dishes. One copy of cells was treated with 0.1 mg/ml hydrocortisone for 24 h as experimental study. The other copy of cells from the same subject was left untreated as paired control. |
| Experimental features | Bone microvascular endothelial cells from femur head of homo sapiens were treated with 0.1 mg/ml hydrocortisone for 24 h as experimental study or left untreated as control. The differential transcriptomes of the two groups were detected and analyzed by using lncRNA–mRNA microarray. The lncRNAs and mRNAs expressed above background were briefly summarized according to their categories (Table 1) or gene ontology enrichments (Fig. 1D). |
| Consent | Allowed for reuse. |
| Sample source location | Beijing, China |
Direct link to deposited data
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60332.
Table 1.
Summary of microarray analysis results.
| Probe class | Total | Expressed above background | Differentially expressed⁎ |
|---|---|---|---|
| lncRNAs | 34,235 | 26,646(78%) | 239(0.9%) |
| mRNAs | 37,585 | 23,833(63%) | 518(2.2%) |
| Combined | 71,820 | 50,479(70%) | 757(1.5%) |
Significant differential expression was defined as probes with P ≤ 0.05 and absolute fold-change ≥ 2.
Experimental design, materials and methods
Ethics statement was approved by the ethics reviewing council of China–Japan Friendship Hospital (Institutional Clinical Trials Register Number is 2014-34, Beijing 100029, China). Written informed consent was obtained from all participating subjects. All patients were enrolled at China–Japan Friendship Hospital between Oct 1, 2013 and Feb 28, 2014, with diagnosis classified as traumatic transcervical fracture. Any traumatic fracture complicated with other etiological mechanisms (e.g. osteonecrosis, arthritis or osteoarthritis) or secondary fracture related to precipitating factors, such as osteoporosis, was excluded.
To harvest bone microvascular endothelial cells (BMECs) for microarray analysis, cancellous bone of femoral head was obtained from orthopedic patients who underwent routine total hip replacement (THR) surgery. The crunched cancellous bones were promptly shipped to the cell culture facility, where they were warmly digested overnight with 0.2% of type one collagenase and subsequent trypsinized of 20 min. After centrifugation, BMECs were collected and plated in 100 mm gelatin-precoated culture dishes, cultured at 37 °C. Complete culture medium was M199 medium supplemented with 2 mM l-glutamine, 100 μg/mL streptomycin and 100 IU/mL penicillin, 20% fetal bovine serum, and recombinant human vascular endothelial growth factor 165. Medium was changed after 24 h to remove nonadherent cells. At day 15, cells were detached by trypsinization and duplicated to 60 mm gelatin-coated culture dishes. After growing confluent, one copy of cells was treated with 0.1 mg/ml hydrocortisone (Tianjin Kingyork Group Co. Ltd., Tianjin, China) for 24 h. The other copy of cells from the same subject left untreated as paired control.
Total RNA was extracted from cells by using the Trizol reagent and purified. The purity and concentration of RNA were determined from OD260/280 readings using spectrophotometer (NanoDrop ND-1000). RNA integrity was determined by capillary electrophoresis using the RNA 6000 Nano Lab-on-a-Chip kit and the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Only RNA extracts with RNA integrity number values > 6 underwent further analysis. Higher yields of cDNA were labeled with a fluorescent dye (Cy5 and Cy3-dCTP) by using CapitalBio cRNA amplification and labeling kit (CapitalBio, Beijing, China). We used two-hybridizations. All controls were labeled with Cy3-dCTP. The experimental samples were labeled with Cy5-dCTP. The labeled cRNAs from lncRNAs and mRNAs were purified and hybridized to Agilent Human lncRNA + mRNA Array V3.0. Images were scanned with the Agilent microarray scanner, gridded, and analyzed using Agilent feature extraction software version 10.10. The raw data were summarized and normalized by using the GeneSpring software V12.0 (Agilent). The mRNAs and lncRNAs expressed above background were hereby selected (Fig. 1A). To select the differentially expressed genes, we used threshold values of ≥ 2 absolute fold change and a Benjamini–Hochberg corrected p value of ≤ 0.05. Student's t-test was used to generate the uncorrected p-values. The data were Log2 transformed and median centered by genes using the Adjust Data function of CLUSTER 3.0 software (Fig. 1B).
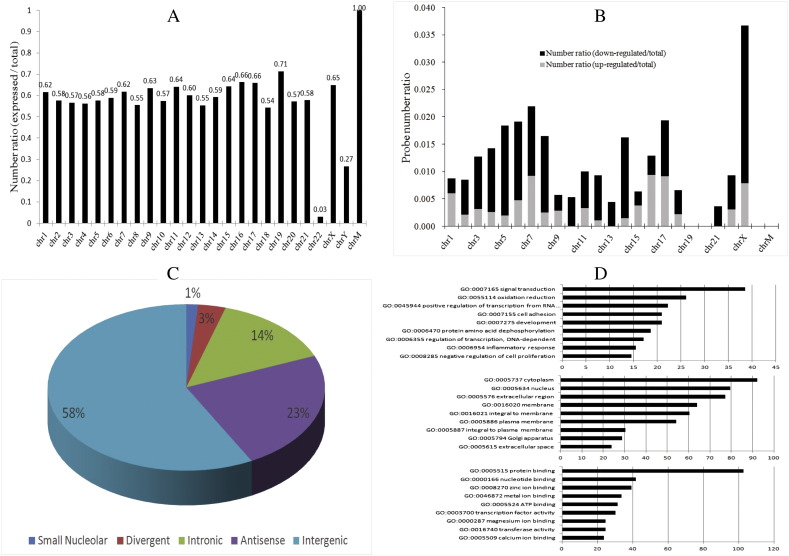
Fig. 1.
General profiles of the differential transcriptomes. (A) Relative distribution of expressed lncRNAs (expressed probes/total probes) derived from each chromosome. (B) Relative chromosomal distribution of significantly up and down-regulated (up or down-regulated probe number/total probes number) lncRNAs. (C) Annotation of genomic context of differentially expressed lncRNAs. Most of the lncRNAs are intergenic, followed by antisense and intronic. No sense overlapping lncRNA is detected. (D) The most enriched terms for differentially expressed mRNAs are displayed here. Enrichment score of the GO equals “− log10 [P-value]”. The listed GO items are categorized into biological process, cellular component or molecular function.
The lncRNAs were classified into five major types: intergenic, intronic, bidirectional (or divergent), sense overlapping and antisense lncRNAs. The annotation we used to perform the lncRNA classification came from the homepage of Arraystar Inc. (http://www.arraystar.com). Besides, a large group of small nuclear ribonucleic acids (snRNAs) are known as small nucleolar RNAs (snoRNAs), which play an essential role in RNA biogenesis and guide chemical modifications of ribosomal RNAs, transfer RNAs and snRNAs (Yin et al., Molecular Cell, 2012). We also consider snoRNAs as a minor type of lncRNA in our study (Fig. 1C). The gene ontolog (GO) enrichment analysis provides a controlled vocabulary to describe differentially expressed transcriptomes. We submitted a list of the 518 statistically differentially expressed mRNAs to MAS 3.0 (http://bioinfo.capitalbio.com/mas3/) for GO term enrichment analysis. The Molecular Annotation System 3.0 (MAS 3.0) is an open platform provided by CapitalBio Corporation. We created a new project and performed the GO analysis following the guidelines (Fig. 1D).
Footnotes
This work was funded by National Natural Science Foundation of China (Reference number 81273972 to Wanshou Guo).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60332.
Table 1.
Summary of microarray analysis results.
| Probe class | Total | Expressed above background | Differentially expressed⁎ |
|---|---|---|---|
| lncRNAs | 34,235 | 26,646(78%) | 239(0.9%) |
| mRNAs | 37,585 | 23,833(63%) | 518(2.2%) |
| Combined | 71,820 | 50,479(70%) | 757(1.5%) |
Significant differential expression was defined as probes with P ≤ 0.05 and absolute fold-change ≥ 2.