Abstract
We report Metagenome from the saline desert soil sample of Little Rann of Kutch, Gujarat State, India. Metagenome consisted of 633,760 sequences with size 141,307,202 bp and 56% G + C content. Metagenome sequence data are available at EBI under EBI Metagenomics database with accession no. ERP005612. Community metagenomics revealed total 1802 species belonged to 43 different phyla with dominating Marinobacter (48.7%) and Halobacterium (4.6%) genus in bacterial and archaeal domain respectively. Remarkably, 18.2% sequences in a poorly characterized group and 4% gene for various stress responses along with versatile presence of commercial enzyme were evident in a functional metagenome analysis.
Keywords: Microbiota, Metagenome, Little Rann of Kutch, Saline desert, Community metagenomics, Functional metagenomics
| Specifications | |
|---|---|
| Organism/cell line/tissue | Saline desert soil metagenome |
| Sex | Not applicable |
| Sequencer or array type | Ion Torrent PGM platform |
| Data format | Raw data: FASTQ file |
| Experimental factors | Environmental sample |
| Experimental features | Whole genome shotgun sequencing followed by community and functional metagenome analysis using MG-RAST online server |
| Consent | Not applicable |
| Sample source location | Saline desert, Little Rann of Kutch, Gujarat State, India |
Direct link to deposited data
https://www.ebi.ac.uk/metagenomics/project/ERP005612
The coding capacity of the soil metagenome is greater than that of the human genome. Metagenomics combines many of the significant molecular technological developments of the last century enabling scientists to investigate more thoroughly microbial ecology and to unlock the vast biotechnological potential held within the microbial population [1]. Besides that phenotype-based screening of bacterial metagenomic libraries provides an avenue for the discovery of novel genes, enzymes, and metabolites that have a variety of potential clinical and industrial uses [2]. Looking for such promising prospects, diverse habitat of worldwide is investigated with metagenomics approach.
The Wild Ass Sanctuary, Little Rann of Kutch, is a typical ecological system with saline desert climate having least floral diversity and exclusive faunal diversity. The sanctuary has been specially protected for the free-ranging population of the Indian Wild Ass (Equus hemionus Khur). The area has dry tropical monsoon climate; it receives an average annual rainfall of less than 300 mm. The average maximum temperature is about 42 °C, and average minimum temperature is about 12 °C. The area is a seasonally flooded wetland ecosystem. During monsoon, a gradual variation of salinity occurs and the concentration gradient of the same is observed from the sea water to the river water [3]. Internationally, the site is recognized for its natural and geomorphologic value and has high conservation value for research and eco-tourism. The transitional climatic conditions existing in the saline desert proposed the possible adaptation of microorganisms in the multistress and multivariable condition of pH, salt, and temperature. On the basis of cultivable approach, microbial community analysis revealed only 20% normophiles with the dominating presence of 40% halophiles and 40% haloalkliphiles [3]. Furthermore, the diazotrophic, IAA and lipase producer microorganism were reported from the same area [4], [5], [6]. Recently the metagenome sequencing of the soil sample from Greater Rann of Kutch was also made available [7]. In present work, we probe the community and functional microbial diversity with metagenomics approach.
Metagenomic DNA was isolated by commercial DNA isolation kit (HiPurA™ Soil DNA kit, MB542; Hi-Media, India) from soil samples collected from the saline desert Little Rann of Kutch, Gujarat, India. Afterward, whole-genome shotgun sequencing was performed using the 318 Chip and 300-bp chemistry Ion Torrent PGM platform as per the manufacturer's instructions. The output was 633,760 sequences with size 141,307,202 bp and 56% G + C content. Subsequently assembly, analysis, and annotation were carried out by MGRAST V 3.5 online server [8].
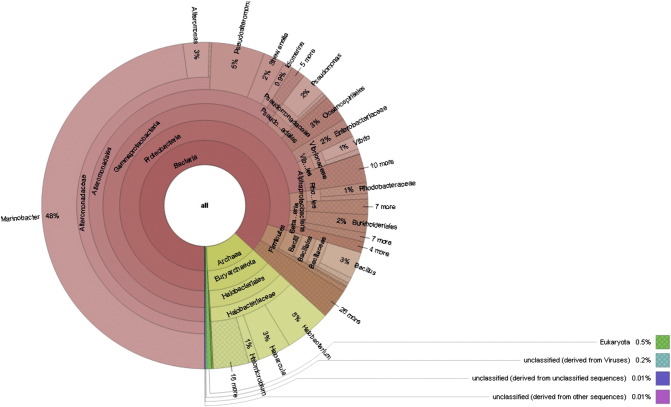
In terms of the kinds of organism present at domain level, 86.6% were bacteria, followed by 12.7% archaea. Moreover, total 43 phyla together with substantial microorganisms in the unclassified category at phyla level were recorded. Likewise, Alteromonadaceae (51.6%) and Halobacteriaceae (12.6%) were major dominating family in bacterial and archaeal domain. Similarly, Marinobacter (48.7%) and Halobacterium (4.6%) were dominating genus for bacterial and archaeal domain. Finally, at species level total 1802 species were reported. The dominating bacterial species were Marinobacter hydrocarbonoclasticus (32.0%) and Marinobacter algicola (12%). While, Halobacterium salinarum (4%) and Haloarcula marismortui (3%) were dominating in archaea (Fig. 1).
Fig. 1.
Community structure of saline desert metagenome.
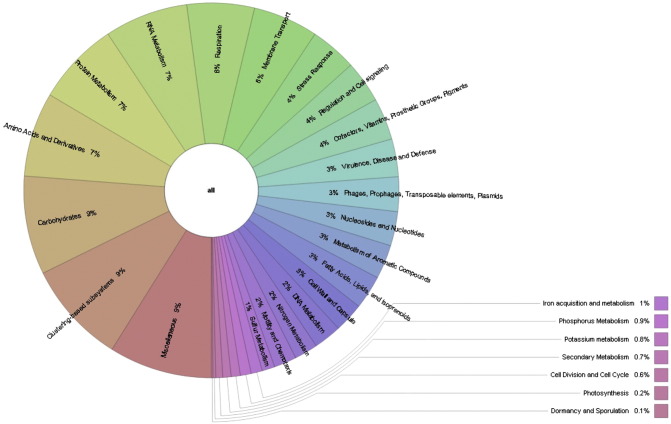
In addition to the community analysis, functional analysis was performed with MGRAST. Out of the 545,855 sequences that passed quality control, 536,652 (98.3%) produced a total of 378,151 predicted protein coding regions. From this 378,151 predicted protein features, 255,915 (67.7% of features) assigned an annotation using at least one of our protein databases (M5NR). Whereas, 122,236 (32.3% of features) have no significant similarities to the protein database. Total 209,895 features (82.0% of annotated features) were assigned to functional categories. In functional annotation with Cluster of Orthologous Group (COG) approach interestingly 56,284 sequences (18.2%) fall in poorly characterized group indicating the possibilities of gaining novel gene from the metagenome. Likewise, in subsystem based annotation approach 4% gene belonged to stress responses for the oxidative stress, osmotic stress, heat shock, cold shock, acid stress, and detoxification. Besides that ample commercial enzyme such as protease, lipases and carbohydratase were also reported in the metagenome (Fig. 2).
Fig. 2.
Functional structure of saline desert metagenome.
Consequently, results suggest rich microbial and gene diversity with possibility of getting novel genes and microorganism for research and commercial application from Little Rann of Kutch, Gujarat, India.
Nucleotide sequence accession number: Metagenome sequence data are available on EMBL-Metagenomics under the accession no. ERP005612.
Acknowledgments
This work was financially supported by Gujarat State Biotechnology Mission (GSBTM) (GSBTM/MD/PROJECTS/SSA/440/2010-2011), Department of Science and Technology (DST), Government of Gujarat, India. We are thankful to Ome Research Facility, Anand Agricultural University, Anand, India for providing the facility of the next generation sequencing.
References
- 1.Steele H.L., Streit W.R. Metagenomics: advances in ecology and biotechnology. FEMS Microbiol. Lett. 2005;247:105–111. doi: 10.1016/j.femsle.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Iqbal H.A., Craig J.W., Brady S.F. Antibacterial enzymes from the functional screening of metagenomic libraries hosted in Ralstonia metallidurans. FEMS Microbiol. Lett. 2014;354:19–26. doi: 10.1111/1574-6968.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koyani R., Patel H., Patel P., Dharaiya N.A., Patel R.K. Study on microbial diversity of wild ass sanctuary, Little Rann of Kutch, Gujarat, India. ICFAI Univ. J. Life Sci. 2009;3:1–8. [Google Scholar]
- 4.Chavada N.B., Patel R.K., Vanpuria S., Raval B.P., Thakkar P.V. A study on isolated diazotrophic (non-symbiotics) bacteria from saline desert soil as a biofertilizer. Int. J. Pharm. Appl. Sci. 2010;1:52–54. [Google Scholar]
- 5.Patel M.V., Patel R.K. Indole-3-acetic acid (IAA) production by endophytic bacteria isolated from saline dessert, the Little Rann of Kutch. CIBTech J. Microbiol. 2014;3:17–28. [Google Scholar]
- 6.Khunt M., Pandhi N. Purification and characterization of Lipase from extreme halophiles isolated from Little Rann of Kutch, Gujarat, India. Int. J. Life Sci. Pharma. Res. 2012;2:55–61. [Google Scholar]
- 7.Pandit A.S., Joshi M.N., Bhargava P., Ayachit G.N., Shaikh I.M., Saiyed Z.M., Saxena A.K., Bagatharia S.B. Metagenomes from the saline desert of Kutch. Genome Announc. 2014;2:2014. doi: 10.1128/genomeA.00439-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer F., Paarmann D., D'Souza M., Olson R., Glass E.M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., Edwards R.A. The metagenomics RAST server — a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinforma. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]