Abstract
Down syndrome (DS, trisomy 21), is the most common viable chromosomal disorder, with an incidence of 1 in 800 live births. Its phenotypic characteristics include intellectual impairment and several other developmental abnormalities, for the majority of which the pathogenetic mechanisms remain unknown. In this “Data in Brief” paper, we sum up the whole genome analysis by mRNA sequencing of normal and DS induced pluripotent stem cells that was recently published by Hibaoui et al. in EMBO molecular medicine.
Keywords: Induced pluripotent stem cells, Down syndrome, Chromosome 21, mRNA sequencing
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens |
| Sex | Female |
| Strain(s) | Induced pluripotent stem cells (iPSCs) from monozygotic twins discordant for trisomy 21: Twin-N-iPSCs versus Twin-DS-iPSCs |
| Sequencer or array type | Illumina HiSeq 2000 instrument |
| Data format | Raw and processed data |
| Experimental factors | Twin-N-iPSCs versus Twin-DS-iPSCs |
| Experimental features | The use of iPSCs derived from monozygotic twins has allowed us to study the effect of the supernumerary chromosome 21 without the biological “noise” that could result from the variability of individual genetic background. The purpose of this RNAseq analysis was to identify differentially expressed genes between Twin-N-iPSCs versus Twin-DS-iPSCs. |
Direct link to deposited data
Deposited data can be found in the Gene Expression Omnibus (GEO) database: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE52251
Experimental design, materials and methods
Cell culture and iPSC derivation
Primary fetal skin fibroblasts were isolated from monozygotic twins discordant for trisomy 21 [1]: Twin-N for the normal fibroblasts and Twin-DS for fibroblasts carrying trisomy 21 anomaly. These samples were obtained post mortem and the study was approved by the ethics committee of the Geneva University Hospital. Normal (Twin-N-iPSCs) and Down syndrome (Twin-DS-iPSCs) induced pluripotent stem cells (iPSCs) were established by transducing the parental fibroblasts (Twin-N and Twin-DS, respectively) with polycistronic lentiviral vectors expressing OCT4, SOX2, KLF4 and c-MYC genes [2], [3]. The generated iPSCs were cultured on human foreskin fibroblasts (ATCC, CCD 1112Sk, Manassas, USA) that were mitotically inactivated by irradiation at 35 Gy before seeding on a gelatin-coated 6-well plate at 3.5 × 105 cells/plate. IPSC colonies were maintained with daily changes in Knock-out Dulbecco's Minimal Essential Medium supplemented with 20% KO serum replacement, 1 mM L-glutamine, 100 μM non-essential amino acids, 100 μM 2-mercaptoethanol, 50 U/mL penicillin and 50 mg/mL streptomycin (all from Gibco, Invitrogen, Basel, Switzerland) and 100 ng/mL human basic fibroblast growth factor (bFGF, Peprotech, London, UK). All iPSC lines were passaged by manual dissection of cell clusters in the presence of 10 μM ROCK-inhibitor Y-27632 (Sigma-Aldrich, Buhs, Switzerland) [4].
RNA extraction and gene expression analysis by mRNA sequencing
Total RNA was extracted from the cell lines, using the QIAGEN (Hilden, Germany) RNeasy MiniKit according to the manufacturer's protocol (Invitrogen). RNA integrity and quantity were assessed with an Agilent (Santa Clara, CA, USA) 2100 bioanalyser, using RNA 6000 nanochips. mRNA-Seq libraries were prepared from 500 ng of total RNA using the Illumina TruSeq™ RNA Sample Preparation kit (Illumina RS-930-2001), following the manufacturer's instructions. Libraries were sequenced on the Illumina HiSeq 2000 instrument to generate 100 bp paired-end reads. Those reads were mapped against the genome (hg 19) using the default parameters of the Burrows–Wheeler Aligner (BWA) [5]. An expression signal was detected in at least one sample for 20456 genes (comprising also non-coding RNA). For each gene, the level of expression was determined by calculating the exon coverage (custom pipeline) and normalizing in Reads Per Kilobase per Million (RPKM). Twin-DS-iPSCs and Twin-N-iPSCs samples were sequenced in three and four biological replicates, respectively, starting from different RNA preparations. Differential expression between Twin-N-iPSCs and Twin-DS-iPSCs was assessed using the default parameters of EdgeR (version 2.4.6) [6] and DESeq (version 1.6.1) [7] programs in R (version 2.14.0). Only the genes with more than 1 read per million in at least 3 replicates were conserved for this analysis. Bonferroni correction was applied to adjust for multiple testing. A gene was considered differentially expressed if the Bonferroni-corrected P-value was lower than 0.01 with both methods. Analysis of the functional annotations associated with the differentially expressed genes was performed using DAVID (Database for Annotation, Visualization and Integrated Discovery) [8], [9].
2. Discussion
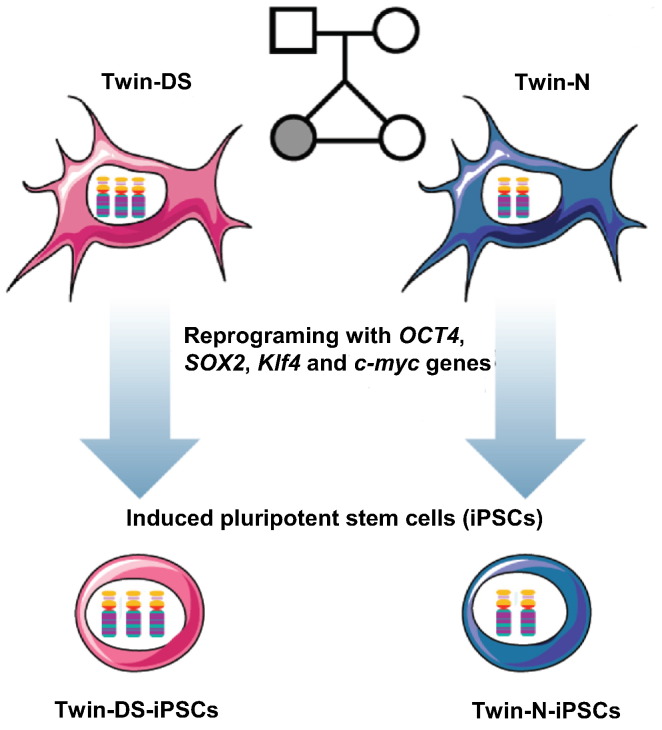
Since the discovery that Down syndrome (DS) is caused by a trisomy of chromosome 21 (HSA21), a major challenge in DS research has been the recapitulation of DS phenotype and the identification of the mechanisms by which the extra copy of HSA21 leads to DS phenotypes [10]. In this respect, disease-specific iPSCs has opened up a new exciting avenue for the modeling and the correction of the phenotypes associated with several neurological diseases (reviewed in [11]). In our recent study, fetal skin fibroblasts isolated from monozygotic twins discordant for trisomy 21 [1], were used to establish normal and DS induced pluripotent stem cells (iPSCs): Twin-N-iPSCs for the normal iPSCs and Twin-DS-iPSCs for the iPSCs carrying trisomy 21 [4] (Fig. 1). As expected for cells that have acquired a pluripotent state, Twin-N-iPSCs and Twin-DS-iPSCs expressed markers of pluripotent cells, showed alkaline phosphatase activity, and differentiated in vitro and in vivo into the three embryonic germ layers mesoderm, ectoderm and endoderm [4].
Fig. 1.
Schematic representation of Twin-N and Twin-DS fibroblast reprogramming into Twin-N-iPSCs and Twin-DS-iPSCs.
Primary fetal skin fibroblasts were isolated from monozygotic twins discordant for trisomy 21 (Twin-N for the normal fibroblasts and Twin-DS for the fibroblasts carrying the trisomy 21 anomaly) and used to establish normal and DS induced pluripotent stem cells (iPSCs) using OCT4, SOX2, KLF4 and c-MYC genes: Twin-N-iPSCs for the normal iPSCs and Twin-DS-iPSCs for the iPSCs carrying trisomy 21 [4].
Then, whole transcriptome analysis was performed using mRNA-Sequencing and the normalized gene expression levels were compared between Twin-N-iPSCs and Twin-DS-iPSCs. Differential expression analysis was performed using the statistical R packages EdgeR and DESeq. Under these conditions, we obtained a list of 624 upregulated and 580 downregulated genes expressed in Twin-DS-iPSCs in comparison with Twin-N-iPSCs (listed in Table 1). The gene ontology (GO) analysis of the 624 genes upregulated in Twin-DS-iPSCs using DAVID showed enrichment for functions predominantly related to the regulation of RNA metabolic processes and regulation of transcription. The GO analysis of the 580 genes downregulated in Twin-DS-iPSCs revealed significant enrichment for genes involved in multiple developmental processes including embryonic development and morphogenesis, organ development and morphogenesis, cellular adhesion and others [4]. Such alterations observed at the iPSC level illustrate the developmental disease transcriptional signature of Down syndrome.
Table 1.
List of the differentially expressed genes between Twin-N-iPSCs and Twin-DS-iPSCs.
Conservative list of 624 upregulated and 580 downregulated genes expressed in Twin-DS-iPSCs in comparison with Twin-N-iPSCs obtained with the statistical R packages EdgeR and DESeq (with a Bonferroni-corrected P-value lower than 0.01).
| Conservative list of 580 downregulated genes expressed in Twin-DS-iPSCs in comparison with Twin-N-iPSCs (with a Bonferroni-corrected P-value lower than 0.01). | Conservative list of 624 upregulated genes expressed in Twin-DS-iPSCs in comparison with Twin-N-iPSCs (with a Bonferroni-corrected P-value lower than 0.01). |
|---|---|
| LOC145845, GPR116, XIST, TSIX, HOXA2, DPP6, FAM181A, MMRN1, SOX1, POU3F2, DMRT2, RXRG, HOXB3, GALP, DLX1, RFX4, HGC6.3, NKX2-3, APLNR, SCRG1, HOTAIRM1, RHOJ, HOXB2, NR2F1, SERPINA3, CLEC1A, LOC440925, SOX17, H19, DKK2, SP5, VCAM1, CLEC3A, SMOC1, CLDN16, WNT1, NTRK2, TBX15, BMP5, CDH5, OTOF, FGF3, MX2, LRRN3, RD3, HABP2, BCHE, EPHA3, EYA4, ISLR2, CDH10, CCDC33, PAX3, LOC283731, DACH1, FLJ31485, COL2A1, BAHCC1, PCDH10, OLIG3, LRRC7, CDH6, RFTN2, CYTL1, SPATA16, ATOH8, ASXL3, DLX2, LGR5, GBX2, MAPK10, SLITRK1, CLEC18B, EOMES, PAX2, ZBTB16, SP8, GLRA1, LOC339535, F13A1, SOX5, RNASE1, AFF3, CCDC48, EGFLAM, UNC5C, GATA4, FZD10, FGF17, NCALD, NCRNA00261, GDF10, MEIS2, CNTFR, NDNF, APOLD1, CDH9, LRRC17, TLR4, EBF2, MGP, MEF2C, ZEB2, FOXA1, SERPINA5, RNF165, GATA6, TAGLN3, FST, GJA5, SLC6A16, ARHGDIB, SH3TC1, LOC149837, FGF9, SULT1C4, FOXA2, C20orf103, ANGPTL1, MTTP, EGR3, NEFM, KCNJ16, SLCO1C1, NKAIN2, ST8SIA2, SOX6, ZNF503, SSPO, EGR2, CDH20, MSX1, SOX9, PPP1R14C, RWDD2B, IGF2AS, TRPC4, RBMS3, WDR52, EPHB3, CXXC4, FBLN7, WDR60, LOC100506385, FZD2, ZNF703, FZD1, CRB2, CX3CL1, TSHZ1, IHH, CD93, GAD1, EFNB1, HNMT, C10orf140, ZNF521, HOGA1, CDH2, CNTN3, CDKN1C, SPOCK3, ADAMTS9, RAI1, LHX1, LOC100130855, MUM1L1, C5, BOC, WLS, KIAA1456, S1PR1, PYGO1, SHOX2, BEAN1, LRP2, LOC284276, THPO, SYNPO, DIO3, RNF212, ST6GALNAC5, EPHA4, USP3, STEAP4, TBX10, FLRT3, DRP2, SCN1A, PAX8, SLC9A9, C1orf213, GLYATL2, HMGCLL1, SLC8A3, ZNF467, F2RL2, IER5L, SULT1A1, SULT1A2, ZNF436, MECOM, LOC646999, TWIST1, SLITRK5, IL2RB, CLDN18, LIX1L, CHRD, TBX20, SLC44A5, SATB2, TMEM121, WNT10B, LOC100505576, IGFBP5, CHRNA9, RBM24, PKNOX2, C15orf2, EDNRB, FGFBP3, LOC144571, FOSB, PDE10A, TRIL, CCDC146, ACCN2, ZNF382, KCNQ1, FIGF, LOC386597, FAM65B, SDK2, KLHL4, GLT25D2, LOC100289178, PREX1, GUSBP1, RIMBP3, TBX5, DOCK10, LHFP, CRABP1, RHOB, LSAMP, NRGN, NRP2, TLR5, NR2F2, ASB9, NFIA, CCDC125, GLDN, TMPRSS5, NCRNA00245, C1QTNF3, COLQ, ZEB1, C6orf118, CHD7, HHAT, SIX1, SCUBE3, DUSP4, SETBP1, SEZ6L, LOC100505881, IQSEC3, BTBD17, VASH1, SORCS2, C5orf38, TMEM169, FLI1, ADCY8, PMFBP1, LYPD1, FRMD4A, RGAG4, MLLT3, SP140L, CHCHD2, C3orf70, DUSP27, PHC2, ZNF518B, PDE4D, MLLT6, MBNL2, FAM38B, C14orf80, NOVA1, PRR4, COL11A1, PRMT8, LGI4, DNAH9, POMZP3, IRX2, DAGLA, MATN3, SDSL, C15orf38, CDH4, PCDHGB6, SRCIN1, SIM2, GYPC, FRMD4B, ONECUT2, STK31, HTRA3, LOC100506190, COL13A1, TMEM132E, EFNB2, FHOD3, CSRNP3, WFIKKN1, CITED1, CAMK2D, PMP22, SYTL2, DENND2A, SOCS3, ZNF704, COLEC12, PRR5L, PDK1, TNFAIP6, DCHS1, PCDHGC4, CUEDC1, FIGNL2, TET2, TBC1D9, PNMA2, GRASP, CTCFL, EFNA5, SNCG, LIMCH1, GSTM3, PCDHGC5, RP9P, LIPG, PCDHGC3, SEMA3A, CRNDE, SLC35F1, KMO, ZADH2, PCDHGA10, PCDHGA9, LRRC27, PCDHGA2, PCDHGA1, BAI1, PCDHGA5, PCDHGA7, PCDHGB1, GFRA2, PCDHGA8, PIK3R1, MGC45800, CPNE8, PCDHGB3, LOC255480, PCDHGA3, EDNRA, PCDHGB2, ARRB1, MXRA5, PCDHGA6, PCDHGB7, ZSWIM6, PCDHGA12, PLEKHA4, BTG2, WIPF1, SOX10, PCDHGA4, IFT140, MDFIC, LOC283624, PCDHGB4, TNRC18, KIAA0101, TRIB2, PCDHGB5, SERTAD4, COL9A2, RNASEL, SYT12, ARC, ID3, ZIC2, IFITM2, LPAR6, ESPNL, HECTD2, H2AFY2, CD99L2, PCDHGA11, NIN, STK32B, SDC2, MN1, ZNF664, COL5A2, SDC3, LPHN2, RNF175, LGALS3BP, SPRY2, CPE, APBA2, ANKRD6, CPXM1, SPRY1, C11orf9, SSBP2, TMSB15A, FZD3, CADM1, NTN1, HTR7, ANTXR2, UHRF1, CCDC167, PCDH18, FAM84A, FKBP7, AADAT, TTC28, BMI1, CDK6, S1PR3, TTC3, PALLD, COMMD3-BMI1, ADAMTS10, KLF12, H2AFY, LMF1, OPN3, TTC3P1, SFRP1, LOC730101, WDR90, FAM115A, MXD4, KIAA1644, FAM89B, CCNG2, ITPKB, TCHP, ST3GAL3, MEX3B, BCL7A, ZNF862, ZNF624, DNAJC15, HEG1, COL4A2, SRRM5, EFS, LCOR, H1F0, MDFI, ALG10B, ENC1, DACT3, SPSB4, SUFU, RNF144A, RNF19A, ANGPTL2, PFKFB4, CD47, GPR153, ITGA4, SEMA5A, PIK3R3, COL9A1, CARD8, ZHX2, ARHGEF40, ELFN1, ZNF280C, C5orf24, GPR161, MCF2L, CAP2, GPSM1, CDK19, CCDC80, GSTA4, PGAP1, KDM6B, CCDC50, ULK2, CCND2, SESTD1, ADAMTS4, SENP7, SMAD3, CSF1, DDHD2, MAP1B, DENND5B, TCF12, YPEL1, PCBP4, SEMA6C, EMILIN1, CPNE2, IGFBP2, MAP7D1, HDAC6, KLF6, JUN, MSI1, ODZ3, APPBP2, FAM176B, C1RL, TMEM123, RGMA, CHD3, MEIS3P1, VIM, FAM113A, MEIS3, CSPG4, PLEKHO1, C6orf89, GPR173, KLF3, DOCK1, NAB1, LOXL2, F2R, ZFP90, SLC35E2, CMTM3, GPX7, LOXL1, LPIN2, SVIL, BTBD6, PDLIM7, TYMS, SHF, ENOSF1, RALGDS, B3GALNT1, GALC, B3GNT5, DOCK7, ZIC5, CDK14, ITGA5, MEGF6, FAM172A, ST5, PAM, STK40, ZMIZ1, EGLN1, JAG1, BNIP3L, UBE2J1, EMID2, BAZ1A, ZNF740, COL4A1, HOOK3, TFDP2. | MTHFD1, SLC1A5, PRR13, KIAA0368, TWISTNB, EIF3B, LIN28A, PODXL, DNAJB6, FOXO1, URB1, TPST2, SALL4, MTL5, CARS, GGCT, AKIRIN1, ACADSB, ZNF134, FAM98A, FAM83G, CLU, ZNF770, VOPP1, RRP12, CDCA7L, PDZD2, GART, SGMS1, IPW, CXADR, ANKRD33B, PFKFB2, CPVL, RAB7L1, PRPS2, ZNF101, MRPS21, RBPMS2, MAD2L2, IDH3A, NME2, KIAA0415, COL9A3, RBBP7, ZNF605, ZNF695, EIF2S3, TBC1D8, PAR-SN, BNC2, TBC1D23, KIAA1543, C21orf59, GCNT2, NFKBIB, C20orf27, C1orf183, PSMA2, MID1IP1, DDX21, CSTB, AARS, MREG, ITPK1, FAM110A, ABCA1, PRKCB, LOC643988, TEX15, CBS, SLC35F2, EDEM2, SESN2, TRIM14, DCN, AEN, DIAPH2, HOOK1, SLC39A14, DPP4, CDS1, ENPP1, B3GNT7, RLTPR, SLC4A5, CLDN6, CCRN4L, PPAP2A, FOXA3, RNF125, ZNF589, MTHFD2, ELOVL4, WIPF3, FBXO41, TBRG4, DRAM1, NUDT6, SPSB2, EIF2S2, PRKAR2B, PHC1, ASTN2, EHHADH, FDXR, USP28, CEBPB, PPARGC1B, PACSIN3, MRPL32, USO1, PPIC, FAM160A1, MRPS17, INADL, MYO1E, EPHA1, WFDC2, CD24, PWP2, GRTP1, SKA3, SYT14, PPM1H, ACOT8, MTA3, DND1, GNAO1, CILP2, KCNS3, GLDC, FGFR4, F11R, C1QTNF1, POLR3G, PSPH, TFAP2C, OLFM2, GREM1, TARS, CCDC69, B3GALT5, CDC42BPG, FOXH1, VWDE, TOMM7, FAM155B, GCH1, TMEM132D, VSNL1, SIRT1, SEMA4A, DPPA4, ZBTB3, ATCAY, FUT1, ALDH1L2, FOSL1, TJP2, SLC44A1, GRM4, ZNF85, TMEM200A, NPTXR, JARID2, GLI1, NACAD, PMAIP1, LLGL2, KCTD14, MBP, FKBP4, ZNF470, HCN2, FBXO27, RBM47, CYCS, POU5F1P3, CHST4, PLS3, RIPK4, SYNGR3, YBX2, SGK1, NFE2L3, PAIP2B, POU5F1B, C8orf44-SGK3, ASNS, SLC7A5, PDK3, LOC100505761, SLC24A2, SLC43A1, GALNT6, ZNF625, CADPS2, MYC, ZNF525, ZNF613, SCAMP5, RYR1, C8orf42, ASS1, ADM2, CCDC3, LOC100506930, ZNF483, DEPTOR, UGP2, FGF2, RAP1GAP2, GNPTAB, PPP2R2C, FZD5, SKIL, SYT13, HPSE, C19orf66, VSIG10, TMEM184A, PRKCQ, RASGRP2, TRIM6, SPR, GARS, CGNL1, UGT3A2, USP44, OSBPL10, CGN, MAFK, UPP1, MRS2P2, MT1X, ZNF398, PHLDA3, INPP5F, NMNAT2, PRSS16, SLCO4C1, ZYG11A, SLC7A3, ZSCAN10, APOE, MYBPC2, MRM1, CCDC64, MRS2, RARRES2, ARHGEF19, NANOG, PDLIM1, KCNC3, SPINT2, CARD11, COL10A1, TMEM38A, SYT3, TXLNG, PPL, ELOVL7, TUBA4A, COMMD7, FRAT2, MFSD6, HERC6, RASIP1, GPR143, FAM101B, ANKRD5, SPINT1, PPP2R2B, SLC1A6, SLC16A10, CLDN9, KSR2, ZNF649, ICOSLG, GPR113, PPP1R16B, GYLTL1B, DOCK9, ZNF165, TC2N, TUBB4, PRKAR1B, IFI30, GRB7, ZNF595, L1TD1, LHFPL4, RNF144B, DENND1C, REEP6, MAP7, PRR5-ARHGAP8, CDH3, CDCP1, EPCAM, C3, LOC100130899, SCNN1A, ARHGAP8, AGPAT9, SGK3, GLB1L3, CDH1, LAMA3, LEFTY1, ELMO3, PLA2G16, PIM2, ARHGEF5, IRF6, GLS2, PALM3, SLC37A1, FAM19A4, JAZF1, C6orf132, LOC728377, SNAI3, SMPDL3B, FAM46B, CYP2S1, COL21A1, RHOV, PROCR, EPS8L2, PPP1R1B, CRB3, SH2D3A, SNCB, STC2, ZDHHC23, C1orf172, KCNH5, LOC339524, AASS, NPW, P2RX2, DHRS3, LAD1, PPAP2C, MAL2, SPP1, SLC27A6, USP43, LOC285484, FBXO2, EREG, HSPA2, RAB19, VSTM2B, AGMAT, KLK1, SLC39A4, TMEM125, TSTD1, LOC157627, RASGRP4, BSPRY, ANKRD24, SLC19A3, GFRA3, PYY, ESRG, FOXD3, GPC4, NOS1, C1orf116, KCNK6, A2ML1, MATK, TRPM6, FERMT1, REPS2, TDGF1, LEFTY2, HEPHL1, NCCRP1, SH3GL2, C1orf88, FGF4, ACAN, GABRD, FAM83F, ZNF296, ITGB1BP3, TERF1, ACP5, CAT, HERC5, NIPAL1, MST1P9, SYT2, STAC2, PNPLA5, PRSS22, KCNK15, ESRP1, LOC643719, TDGF1P3, PCP4L1, ACE, SLC2A14, CHST8, TRIB3, ICA1, C1orf210, C16orf11, GPR160, CLDN7, BEND4, EHF, CTAGE4, TRIM58, TNFRSF8, CCNI2, ST14, ZNF860, ECHDC2, MMP3, MACC1, SFRP5, LOC100506428, LPAR3, RIMS4, CBR3, OVOL1, SLC30A3, BHMT2, HEY2, BCAN, MCOLN2, SLC4A11, VAV1, BHMT, INHBE, KLRG2, ZNF850, CHAC1, ZDHHC22, EPO, TNNC2, ZMYND15, CA4, CHD5, COBL, ST8SIA5, C20orf118, GPR172B, RTBDN, ZNF513, CTAGE15P, CBR1, MMP24, LOC100133286, MARVELD3, PTPRN, ZNF876P, CCDC64B, GPR64, BBS9, ESRP2, RBP7, AIM1, UTF1, NAPRT1, FBXL21, SAMHD1, CABP1, TMEM151A, CR1L, HESX1, C2orf65, HES2, EGFL6, ILDR1, CTSF, RPL39L, HMX1, ZNF493, TFCP2L1, CNTN1, PCDHB17, PKP3, LOC730102, CXCL16, MYH2, LOC100288748, DNMT3B, CCNA1, CR2, NRN1L, FA2H, TULP2, PROKR2, C1orf130, ANKRD34B, KLK13, C17orf107, HRASLS5, AP1M2, EVC2, SOX15, ADAD2, LYPD5, LOC647946, LOC284408, TMEM215, MFSD6L, LOC100631378, PCDHB15, DPPA2, TRDN, ZNF354C, SLFN12, SCIN, LOC84856, NLRP7, WIF1, MYOZ1, CSF3, NLRP2, ZNF441, SCNN1G, C8orf47, HRH3, ZNF680, ALPK3, ZNF506, ZNF471, GABRG1, MGMT, FLJ34208, ZNF799, ZNF585B, ERAS, ZNF440, ZNF443, ZNF229, ZNF528, ZNF454, CLEC4GP1, SCNN1B, ZNF630, ZFP28, AGBL2, ZNF578, ZNF662, NR1I2, NNAT, CLEC4G, RAMP3, PCDHB5, TCL1A, ZNF682, PIWIL2, LOC100128252, NAP1L6, KLHL34, LOC440910, ZNF433, ZNF135, ZNF823, ZNF829, EPHX3, ZNF667, ZNF90, DPPA3, ZNF563, ZNF439, ZNF44, ZNF572, ZNF442, ZNF69, ZNF502, ZNF429, LOC644554, RBM46, ZNF844, ZNF763, ZNF737, ZNF700, MYH14, DPPA5, ZNF732, CST6, ZNF253, NUDT16P1, GPR114, TMEM30B, ZNF826P, ZNF835, ZNF736, ZNF486, ZNF518A, ZNF626, ZSCAN1, LOC399815, MARVELD2, ZNF560, LY75, ZNF788, MT1H, FOXI2, PEG3, ZNF492, ZNF93, CYP4F22, ZNF98, DCAF12L1, ZNF257, LOC441666, LOC400680, LOC400655, ZNF208, ZFP42, ZNF878, NLRP4, ZNF99. |
Considering our results and the recent studies recapitulating and rescuing the phenotypes associated with several neurological diseases using pluripotent stem cell-based models (reviewed in [11]), we believe that a combination of the transcriptional profiling with the possibility to derive in principle all cell type relevant for the disease will contribute to a better understanding of the initiation and the development of DS and ultimately to the design of new therapies.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
This work was supported by grants from the Ernest Boninchi Fondation to A.F., Genico to A.F., the Swiss National Science Foundation (SNF-144082) and the European Research Council (ERC-249968) to S.E.A.
Contributor Information
Stylianos E. Antonarakis, Email: Stylianos.Antonarakis@unige.ch.
Anis Feki, Email: Anis.Feki@h-fr.ch.
References
- 1.Dahoun S. Monozygotic twins discordant for trisomy 21 and maternal 21q inheritance: a complex series of events. Am. J. Med. Genet. A. 2008;146A(16):2086–2093. doi: 10.1002/ajmg.a.32431. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Grad I. NANOG priming before full reprogramming may generate germ cell tumours. Eur. Cell Mater. 2011;22:258–274. doi: 10.22203/ecm.v022a20. [DOI] [PubMed] [Google Scholar]
- 4.Hibaoui Y. Modelling and rescuing neurodevelopmental defect of Down syndrome using induced pluripotent stem cells from monozygotic twins discordant for trisomy 21. EMBO Mol. Med. 2014;6(2):259–277. doi: 10.1002/emmm.201302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 9.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonarakis S.E. Chromosome 21 and Down syndrome: from genomics to pathophysiology. Nat. Rev. Genet. 2004;5(10):725–738. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- 11.Hibaoui Y., Feki A. Human pluripotent stem cells: applications and challenges in neurological diseases. Front. Physiol. 2012;3:267. doi: 10.3389/fphys.2012.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]