Abstract
FOXO family members (FOXOs: FOXO1, FOXO3, FOXO4 and FOXO6) are important transcription factors and tumor suppressors controlling cell homeostasis and cell fate. They are characterized by an extraordinary functional diversity, being involved in regulation of cell cycle, proliferation, apoptosis, DNA damage response, oxidative detoxification, cell differentiation and stem cell maintenance, cell metabolism, angiogenesis, cardiac and other organ’s development, aging, and other critical cellular processes. FOXOs are tightly regulated by reversible phosphorylation, ubiquitination, acetylation and methylation. Interestingly, the known kinases phosphorylate only a small percentage of the known or predicted FOXOs phosphorylation sites, suggesting that additional kinases that phosphorylate and control FOXOs activity exist. In order to identify novel regulators of FOXO3, we have employed a proteomics screening strategy. Using HeLa cancer cell line and a Tandem Affinity Purification followed by Mass Spectrometry analysis, we identified several proteins as binding partners of FOXO3. Noteworthy, Polo Like Kinase 1 (PLK1) proto-oncogene was one of the identified FOXO3 binding partners. PLK1 plays a critical role during cell cycle (G2-M transition and all phases of mitosis) and in maintenance of genomic stability. Our experimental results presented in this manuscript demonstrate that FOXO3 and PLK1 exist in a molecular complex through most of the phases of the cell cycle, with a higher occurrence in the G2-M cell cycle phases. PLK1 induces translocation of FOXO3 from the nucleus to the cytoplasm and suppresses FOXO3 activity, measured by the decrease in the pro-apoptotic Bim protein levels and in the cell cycle inhibitor protein p27. Furthermore, PLK1 can directly phosphorylate FOXO3 in an in vitro kinase assay. These results present the discovery of PLK1 proto-oncogene as a binding partner and a negative regulator of FOXO3 tumor suppressor.
Keywords: FOXO tumor suppressors, transcription factors, Polo Like Kinase 1, proto-oncogene, FOXO3 binding partners, FOXO1, PI3K-Akt pathway, apoptosis, cell cycle
INTRODUCTION
FOXO family members (FOXO1, FOXO3, FOXO4 and FOXO6) are important transcription factors and tumor suppressors involved in the control of cell homeostasis and cell fate. They regulate various critical functions, having a “remarkable functional diversity”1,2, regulating cell cycle, proliferation, apoptosis, DNA damage response, oxidative detoxification, cell differentiation and stem cell maintenance, cell metabolism, angiogenesis, cardiac development, aging and other cellular processes2.
-
FOXO family members are important transcription factors involved in the control of critical and very divers cellular functions and processes, such as:
cell cycle, proliferation, apoptosis, DNA damage response, oxidative detoxification, cell differentiation and stem cell maintenance, cell metabolism, angiogenesis, cardiac development, aging, and other critical cellular processes
FOXO3 activation can result in modulation of multiple critical cellular processes, such as cell cycle arrest (through upregulation of p27, p21 expression, and downregulation in Cyclin D1 and Cyclin D2 expression)2 and apoptosis. FOXO-induced apoptosis can be initiated by both the intrinsic pathway (upregulation of Bim, Puma, Noxa) and extrinsic pathway of apoptosis (upregulation of Apo2L/TRAIL, FasL and TRADD)2–6. Immunometabolism is also a new cutting edge area of research that investigates the bidirectional regulation between immune system and metabolism7. FOXOs and PI3K pathway are critical in regulation of both the immune system8,9 and the metabolism10.
FOXO family members (FOXOs) are tightly regulated by reversible phosphorylation, ubiquitination, acetylation and methylation1,2,11. FOXOs can be inactivated by direct phosphorylation of several well known kinases, such as AKT, IKK, SGK and ERK, resulting in their nuclear exclusion and transcriptional inhibition2,12–14. Our previous studies identified PP2A as the first phosphatase that binds and dephosphorylates FOXO3 at AKT-dependent sites, thus reactivating the FOXO3 transcription factor15. On the other hand, phosphorylation of other FOXOs phosphorylation sites by few specific kinases can result in its cytoplasmic – nuclear translocation and activation. For example, MST1 phosphorylates FOXO3 at Ser207, which induces FOXO3 dissociation from the scaffold protein 14-3-3, causing relocalization to the nucleus and transcriptional-dependent expression of FOXO3 targets, such as the pro-apoptotic Bim15–17. Interestingly, the known kinases phosphorylate only a small percentage of the known or predicted FOXOs phosphorylation sites18. This suggests that additional kinases that phosphorylate and control FOXOs activity exist.
The inhibition of FOXO tumor suppressors transcriptional activity, which can be done by several mechanisms, including AKT or ERK-mediated inactivation or mutations (fusion mutants), is an important mechanism identified in a variety of malignancies19–21. For example, Bcr-Abl mediated suppression of FOXOs was recently shown to be critical for progression of Leukemia,2,12,22,23. Thus, the discovery of novel mechanisms and strategies that regulate FOXOs and can be targeted for re-activation of FOXOs hold promise for cancer treatment.
In order to identify novel regulators of FOXO3, we have employed a proteomics screening strategy. Using HeLa cancer cell line and a Tandem Affinity Purification followed by Mass Spectrometry analysis, we identified several proteins as novel binding partners of FOXO3. Noteworthy, Polo Like Kinase 1 (PLK1) proto-oncogene was identified as a FOXO3 binding partner. PLK1 is a member of the Polo-Like kinases family, comprising of PLK1, PLK2 and PLK3, a conserved family of kinases, critical in regulation of cell cycle/proliferation and in DNA damage-induced checkpoints24.
PLK1 plays a critical role during mitosis and in maintenance of genomic stability. PLK1 is a proto-oncogene overexpressed in a wide range of malignancies (including but not limited to melanoma, non-small lung cancer, gastroinstestinal cancers and prostate cancer). Interestingly, PLK1 is associated with an increased risk for metastasis24–27. Several PLK1 inhibitors have been developed and PLK1 suppression with small molecules is analyzed in clinical trials for the treatment of Acute Myelocytic Leukemia (AML) and other malignancies23,28–31.
Our experimental results presented here demonstrate that FOXO3 and PLK1 exist in a molecular complex throughout most of the phases of the cell cycle, with a higher occurrence in the G2 and M phases of the cell cycle. PLK1 induces translocation of FOXO3 from nucleus to the cytoplasm and suppresses FOXO3 activity, measured by the decrease in the protein levels of the pro-apoptotic Bim and of the cell cycle inhibitory protein p27. Furthermore, PLK1 can directly phosphorylate FOXO3 in an in vitro kinase assay. These results present the discovery of PLK1 as a binding partner of FOXO3 that negatively regulates FOXO3 localization and activity.
MATERIALS AND METHODS
Cell lines, transfections and synchronization
HeLa and HEK293T cell lines were obtained from ATCC (American Type Culture Collection) and maintained in Dulbecco’s modified Eagle’s Medium (DMEM; Mediatech Inc., Manassas, VA, USA) supplemented with 10 % Fetal Bovine Serum (FBS, HyClone/Thermo Scientific, Waltham, MA, USA), L-Glutamine (Mediatech Inc.) and Penicillin/Streptomycin (Mediatech Inc.) (complete DMEM). All cell lines were cultured in an atmosphere of 37C and 5% CO2.
Transient transfections of DNA plasmids were done by using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA), as described before32,33, according to the manufacturer’s specifications. For experiments involving co-transfection, total transfected DNA was held constant by the addition of an empty control plasmid (pcDNA3).
When specified, cells were synchronized with Nocodazole for 24h and released. Cells collected at 0, 5, 10, 15 and 20h after Nocodazole release were evaluated for cell cycle phases by analyzing the Cyclins expression (B1, A), which are particularly expressed in specific phases of the cell cycle and PLK1 (mainly expressed in G2 and M phases of the cell cycle)34.
Plasmids
pcDNA3-FLAG-HA plasmid was provided by William Sellers (DFCI, Harvard Medical School)35. pcDNA3-FLAG-HA-FOXO3 and TM (FOXO3 triple mutant with T32, S253, and S315 modified to Alanine) were developed by PCR cloning. pcDNA3-FLAG-HA and pcDNA3-FLAG-HA-FOXO3 were used in the proteomic screening. FOXO3 mutations were generated by standard PCR based site-directed mutagenesis (Stratagene) using pcDNA3-FLAG-HA-FOXO3 as a template. pcDNA3-FLAG-FOXO1 was provided by Kun-Liang Guan (Moores Cancer Center, University of California from San Diego, CA, USA)36. FOXO1 mutants were generated by standard PCR based site-directed mutagenesis. GLOFLAG3-FLAG-FOXO4 was provided by Boudewijn Burgering (University Medical Center Utrecht, Utrecht, Netherlands)37. GST-FOXO3 was bought from Addgene (GST-FOXO3a WT, Plasmid #1790). pcDNA3-PLK1 plasmid was provided by Professor Wenyi Wey (BIDMC, Harvard Medical School, Boston, USA). The shRNAs were provided by Sigma. Purified PLK1 kinase was purchased from Cell Signaling.
Western Blot
Western Blot was performed as described before38,39. Briefly, the cells (10 cm dishes) were washed twice with phosphate-buffered saline (PBS) then scraped on ice in either RIPA buffer (Boston BioProducts), or EBC lysis buffer (in immunoprecipitation experiments) (50mm Tris-HCL [pH 8.0], 120mM NaCl, 0.5% [v/v] Nonidet P-40 (NP-40), and 5mM EDTA) supplemented with protease inhibitors (Complete, Roche Applied Science) and phosphatase inhibitors (Halt Phosphatase Inhibitor Cocktail, Pierce Biotechnology). Protein concentrations were measured by using the BCA protein assay reagent (Pierce). Equal amounts of soluble protein were diluted with EBC or RIPA lysis buffers, followed by SDS-PAGE, transfer and probing with the specified antibodies.
Antibodies
The following antibodies were purchased from Cell Signaling Technology: PLK1 (208G4), pan-Akt (C67E7), (C31E5), pAkt (S473) (193H12), FOXO3 (75D8), and Bim (#2819). The following antibodies were purchased from Santa Cruz Biotechnology: pan 14-3-3 (K-19), FOXO3 (H-144). Anti-FOXO3a/FKHRL1 Antibody (07-702) was purchased from Millipore. The FLAG (M2)-horseradish peroxidase (HRP) conjugate and β-actin (clone AC-15) were purchased from Sigma. For immunoblotting, all antibodies were used at a 1:1 000 dilution with the exception of the anti-FLAG (M2)-HRP (1:20 000), anti-β-Actin (clone AC-15) (1:20 000) and anti-pan 14-3-3 (1:2 000).
Immunoprecipitation/Co-immunoprecipitation
For immunoprecipitation15, cells were lysed in EBC lysis buffer and the whole cell lysates were then pre-cleared for 1h at 4°C with protein A/protein G plus agarose (Calbiochem) and then incubated for 2h at 4°C while rotating with anti-FLAG (M2) affinity gel (Sigma) (20μL packed beads). FLAG immunocomplexes were washed three times with EBC buffer supplemented with protease and phosphatase inhibitors before being boiled in Laemmli reducing sample buffer. Immunocomplexes and 5%–10% of the immunoprecipitation input from the initial whole cell lysate were analyzed by immunoblotting as indicated in the respective figures. To detect the endogenous interactions, whole cell lysate (2–3mg) was pre-cleared for 1h at 4°C with protein A/protein G plus agarose and then incubated overnight at 4°C while rotating with either normal rabbit immunoglobulin G (IgG) (Santa Cruz Biotechnology) or anti-FOXO3 antibody (H-144; Santa Cruz Biotechnology) (1μg IgG/mg lysate). Immunocomplexes were then captured with protein A/protein G plus agarose (20μL packed beads/mg of lysate) for 1h at 4°C and processed as described above. Endogenous immunoprecipitated FOXO3 was detected by immunoblotting with an anti-FOXO3 rabbit antibody (Cell Signaling).
Tandem affinity purification (TAP) and mass spectrometry15
Briefly, HeLa cells expressing exogenous FOXO3 fused to tandem FLAG and HA epitopes (FOXO3-FLAG-HA) were lysed in EBC buffer as described above. Cell lysates were first immunoprecipitated with anti-FLAG (M2) affinity gel (Sigma) for 2h at 4°C.
FLAG immunocomplexes were washed three times with EBC lysis buffer and FOXO3-FLAG-HA was eluted with 150ng/μl 3X FLAG peptide (Sigma). FLAG peptide eluates were then immunoprecipitated with anti-HA (HA7) agarose affinity resin (Sigma) for 2h at 4°C, washed three times with EBC buffer, and eluted with 1μg/μl HA peptide (Sigma). The lysate was then fractionated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then silver stained. The marked protein bands were excised from the gel and analyzed by microcapillary LC/MS/MS techniques (Taplin Mass Spectrometry Core Facility, Harvard Medical School, Boston, MA, USA) by using a LTQ FT Ultra Hybrid Mass Spectrometer (Thermo Electron).
RESULTS
Novel FOXO3 binding partners discovered by using a TAP-MS strategy
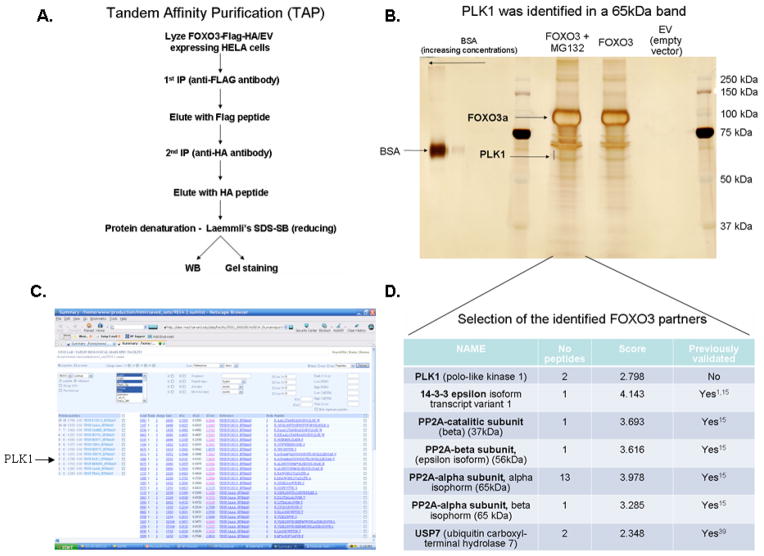
In order to discover novel binding partners and regulators of FOXO3, we have employed a Tandem Affinity Purification (Figure 1A). Co-immunoprecipitated proteins with FOXO3 were separated by SDS-PAGE, stained with Silver stain and the binding partners were identified by Mass Spectrometry (Figure 1B). Interestingly, one of the discovered proteins is PLK1 (Figure 1B, C & D), a proto-oncogene kinase critical for cell cycle progression and important in tumorigenesis and metastasis of various types of cancers23–29. Moreover, other binding partners were also identified in complex with FOXO3. A selection of some of these proteins is presented in Figure 1D. Several of the PP2A subunits were also identified. We have previously reported PP2A as a phosphatase that can dephosphorylate FOXO3 at AKT-dependent phosphorylation sites leading to its activation15. Moreover, several known FOXOs binders were also identified in our screening, which validates our strategy. For example, USP7 was previously identified as a FOXO4 deubiquitinating enzyme40, while 14-3-3 is a well known scaffold protein that binds and keeps FOXOs in the cytoplasm, where they can be degraded41.
Figure 1. Novel FOXO3 binding partners discovered by using a TAP-MS strategy.
A. Tandem Affinity Purification (TAP) followed by protein identification by Mass Spectrometry (MS) method was employed to identify novel FOXO3 binding partners. Lysates from HeLa cells transfected with either FOXO3-Flag-HA or EV-Flag-HA were immunoprecipitated with an anti-FLAG antibody, followed by a second purification with an anti-HA antibody. Denatured proteins (reducing Laemmli’s SDS-SB) were separated by SDS-PAGE. B. Proteins were stained with Silver stain. The bands corresponding to FOXO3 and PLK1 proteins identified by MS are presented. BSA (Bovine Serum Albumin) was used as a marker for protein quantity. C. Proteins identified by MS in the 60–65 kDa band presented in (B). PLK1 was one of the identified proteins (the arrow indicates PLK1). D. Most important/promising proteins identified after analyzing multiple bands by MS, covering 25–200 kDa gel range, are presented. The “Score” represents a probability Score determine with Sequest. EV – empty vector; MG132 was used to enrich for potential E3 ligases.
Screening validation: PLK1 kinase forms a complex with FOXO3 and other FOXO family members (FOXO1 & FOXO4)
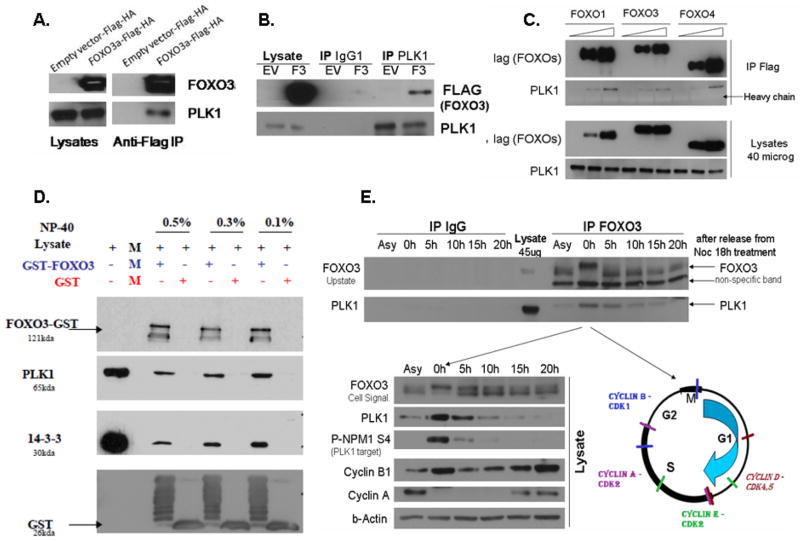
We next validated the PLK1-FOXO3 interaction by using a co-immunoprecipitation strategy in HeLa cells. Exogenous FOXO3 co-immunoprecipitates with endogenous PLK1 (Figure 2A), while endogenous PLK1 also co-immunoprecipitates with exogenous FOXO3 (revers co-immunoprecipitation, Figure 2B). Since in many cellular settings FOXO family members have redundant functions, we aimed to determine if other FOXO family members also form a complex with PLK1. Indeed, exogenous FOXO1 and FOXO4 form a complex with endogenous PLK1, suggesting that FOXO-PLK1 interaction is conserved between FOXO family members (FOXO1, FOXO3 and FOXO4). This suggests that a conserved sequence in FOXO3 is responsible for PLK1 binding (Figure 2C).
Figure 2. Screening validation: PLK1 kinase forms a complex with FOXO3 and other FOXO family members (FOXO1 & FOXO4).
A. HeLa cells were transfected with either pcDNA3-EV-FLAG-HA (Control) or pcDNA3-FOXO3-FLAG-HA and exogenous FOXO3 was immunoprecipitated by using an anti-FLAG antibody. The results show that exogenous FOXO3 is co-immunoprecipitated with endogenous PLK1 in HeLa cells. B. HeLa cells were transfected with either pcDNA3-EV-FLAG-HA (Control) or pcDNA3-FOXO3-FLAG-HA and endogenous PLK1 was immunoprecipitated by using an anti-PLK1 antibody. The results show that endogenous PLK1 is co-immunoprecipitated with exogenous FOXO3 in HeLa cells (revers immunoprecipitation). C. HeLa cells were transfected with either pcDNA3-EV-FLAG (Control), pcDNA3-FOXO1-FLAG, pcDNA3-FOXO3-FLAG and pcDNA3-FOXO4-FLAG. Exogenous FOXO1/FOXO3/FOXO4 was immunoprecipitated by using an anti-FLAG antibody. The results demonstrate that all three FOXO family members can form a complex with endogenous PLK1; D. Purified GST-FOXO3 protein was incubated with lysates from PLK1-FLAG transfected HeLa cells. GST pull-down reveals that GST-FOXO3 forms a complex with PLK1 (detected with an anti-FLAG antibody). GST is used as a negative control, while 14-3-3, a known scaffold protein of FOXO3, is used as a positive control. Results obtained with several different concentrations of NP40 detergent are shown. E. Asynchronous and synchronized proliferating HeLa cells in different phases of the cell cycle were used to evaluate the endogenous-endogenous interaction between FOXO3 and PLK1. Cells were synchronized with Nocodazole and released. Cells collected at 0, 5, 10, 15 and 20h after Nocodazole release were evaluated for cell cycle phases by analyzing the Cyclins expression (B1, A), which are particularly expressed in specific phases of the cell cycle (see right panel of the lower figure). PLK1 is also expressed at highest levels in mitosis and can itself be used as an indicator of cell cycle phase. Immunoprecipitated endogenous FOXO3 co-immunoprecipitates with endogenous PLK1. While the FOXO3-PLK1 complex can be seen during all phases of the cells cycle and in asynchronous cells, most PLK1 is bound to FOXO3 during the M/G2-M phases of the cell cycle. This is probably due to the fact that PLK1 has the highest expression during these phases of the cell cycle; PLK1 activity is determined by measuring the phosphorylation of NPM at Ser4 (site phosphorylated by PLK1)42.
We further validated the PLK1-FOXO3 interaction by using GST pull-down studies. Purified GST-FOXO3 forms a complex with exogenous PLK1 (detected with an anti-FLAG antibody) (Figure 2D). Moreover, endogenous FOXO3 also co-immunoprecipitates with endogenous PLK1 (Figure 2E). While FOXO3-PLK1 endogenous-endogenous interaction occurs in almost all phases of the cell cycle, the highest amount of FOXO3-PLK1 complexes can be seen during the M/G2-M phases of the cell cycle (Figure 2E), when the expression of PLK1 is the highest. PLK1 is critical for G2-M transition, M phase progression and M-G1 phase transition. Thus, its highest expression occurs during these phases of the cell cycle23,26, as also observed in Figure 2E (lower panel). Interestingly, there is a shift in the FOXO3 band in the M/G2-M phases of cell cycle (Figure 2E, upper panel), which may be due to the mitosis-induced post-translational modifications of FOXO3, such as phosphorylation by mitosis-activated kinases. Further experiments need to be performed to confirm this hypothesis.
We also confirmed the endogenous-endogenous interaction in 293T cell line (Supplementary Figure 1). PLK1 activity is determined by measuring the phosphorylation of NPM at Ser4 (site phosphorylated by PLK1)42. Cyclins were used to estimate the cell cycle phases.
PLK1 binds FOXO3 at specific sites
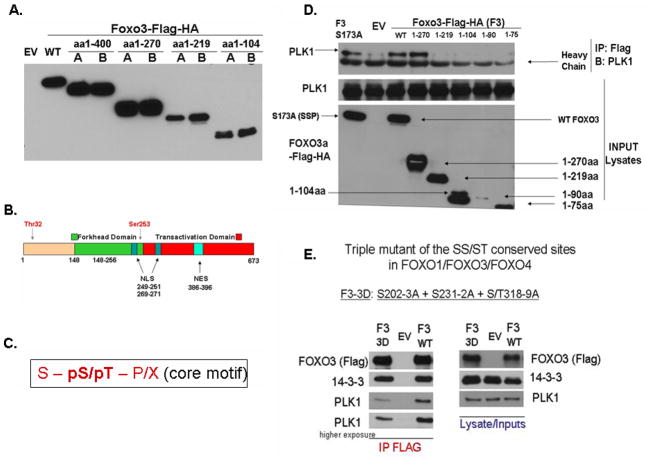
To get more insights into what region or specific sites in FOXO3 bind PLK1, we designed and developed several FOXO3 deletion mutants, corresponding to the amino acids (aa) 1-400, 1-270, 1-219, 1-104 (Figure 3A), which may include one or more functional domains of FOXO3 (Figure 3B). Moreover, we also designed mutants of the FOXO3 and FOXO1 core consensus binding motifs43,44 predicted to bind PLK1. One core consensus binding motif (S-pS/pT-P/X, Figure 3C, where “S” is Serine, “pS” is phosphorylated Serine, “pT” is phosphorylated Threonine and “P” is Proline) exist in FOXO3 (S173), three in FOXO1 (S41, S383, S394) and none in FOXO4 (Supplementary Figure 2). We designed mutants (site directed mutagenesis) for all these sites (S173A (FOXO3), S41A, S383A and S394A and combinations of these (FOXO1)). Our results demonstrate that PLK1 can interact with aa 1-270, but not with aa 1-219 of FOXO3, suggesting that the region between aa 219–270 is important for binding (Figure 3D). Mutations of the core consensus sequences in FOXO3 and FOXO1 do not have any effect on PLK1 binding to FOXO3 (Figure 3D (FOXO3) and data not shown (FOXO1)). This is what we actually expected, since the core consensus sequence is not preserved between FOXO1, FOXO3 and FOXO4 and we previously shown that PLK1 binds to all these three FOXO family members. However, if we consider a shorter variant of the core consensus sequence (S-pS/S-pT), there are three sites that are conserved between the three FOXO family members. A mutant of all these three sites significantly decreases the binding of PLK1 to FOXO3, suggesting that one or more of these sites are important in binding (Figure 3E). As expected, one of these three sites (S231-S232) is found in the 219–270 region, which we have established to be important in binding, suggesting that this site may have a significant contribution to the PLK1 binding to FOXO3. However, single mutants do not seem to have a striking effect on decreasing PLK1-FOXO3 binding (data not shown). Also, the triple mutant does not completely abolish the PLK1 binding to FOXO3, suggesting that additional binding sites may exist. 14-3-3 similarly binds FOXO3 (F3 WT) and the triple mutant of FOXO3 (F3 3D), suggesting that the decrease in binding is not due to a non-specific triple mutation-induced disruption of FOXO3 conformation.
Figure 3. PLK1 binds FOXO3 at specific sites.
A. Testing of the FOXO3 C-terminal deletion mutants – size dependent SDS-PAGE migration - HeLa cells were transfected with either pcDNA3-EV (Control) or pcDNA3-FOXO3 deletion mutants and detected with an anti-FLAG antibody. Constructs were validated by checking both the correlation between their size and migration on SDS-PAGE, and sequencing of each individual construct. Two different clones were selected after the mutagenesis process and named A and B. Clone A was further selected for the following experiments; B. Schematic structure of FOXO3 protein underscoring its major functional domains (reproduced with permission2). C. PLK1 PBD conserved core consensus sequence as determined by Lewis C. Cantley & Michael B. Yaffe43,44 (“S” is Serine, “pS” is phosphorylated Serine, “pT” is phosphorylated Threonine and “P” is Proline); D. Finding PLK1 binding region in FOXO3. HeLa cells were transfected with either pcDNA3-EV (Control) or pcDNA3-FOXO3 deletion mutants and exogenous FOXO mutants were co-immunoprecipitated (using an anti-FLAG antibody) with endogenous PLK1, in HeLa cells. Results show that the region between the amino acids (aa) 219–270 of FOXO3 is important for PLK1 binding; E. A mutant of the three SS/ST conserved sites between FOXO family members significantly suppresses the binding of PLK1 to FOXO3.
PLK1 regulates FOXO3 nuclear localization and total levels
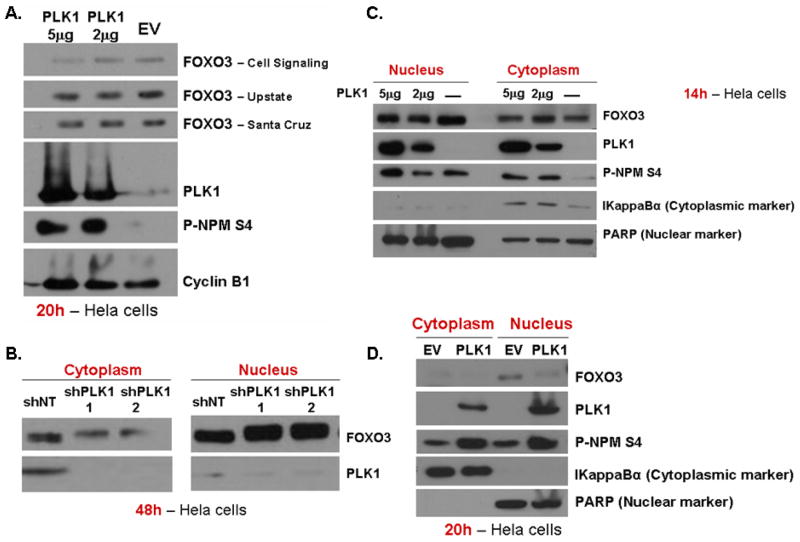
In order to establish if PLK1 has any effect on FOXO3 total levels, increasing levels of PLK1 protein were expressed in HeLa cells (Figure 4A). PLK1 expression induces a decrease in total levels of FOXO3 (Figure 4A). We further investigated if PLK1 knock-down (Figure 4B) or PLK1 expression (Figure 4C & D) have any effect on FOXO3 localization. Endogenous PLK1 knock-down induces endogenous FOXO3 localization into the nucleus (Figure 4B), while PLK1 expression decreases endogenous FOXO3 levels in the nucleus, showing that FOXO3 localization is controlled by PLK1. FOXOs localization into the nucleus is a requirement for its transcriptional activity, since it needs to bind the DNA45,46. Thus, these results suggest that PLK1 induces the exclusion of FOXO3 from the nucleus, which should result in the inhibition of its transcriptional functions.
Figure 4. PLK1 regulates FOXO3 nuclear localization and total levels.
A. HeLa cells were transfected with either EV-FLAG or increasing concentrations (2ug, 5ug) of PLK1-FLAG. FOXO3 total levels are downregulated by PLK1 overexpression after 20h from transfection. Cyclin B1 is used as an internal loading control. B. HeLa cells were transfected with either two different shRNA for PLK1 or a non-targeting shRNA (shNT), for 48h. PLK1 knockdown induces FOXO3 relocalization into the nucleus. Nuclei and cytoplasm were isolated and FOXO3 levels were evaluated in each fraction. C. & D. HeLa cells were transfected with either EV-FLAG or PLK1-FLAG. PLK1 expression induces a decrease of the nuclear FOXO3 fraction at both 14h (C.) and 20h (D.), suggesting a decrease in FOXO3 activity. PARP is used as a nuclear marker, while IkappaBα is employed as a cytoplasmic marker; PLK1 activity is determined by measuring the phosphorylation of NPM at Ser4 (site known to be phosphorylated by PLK1)42;
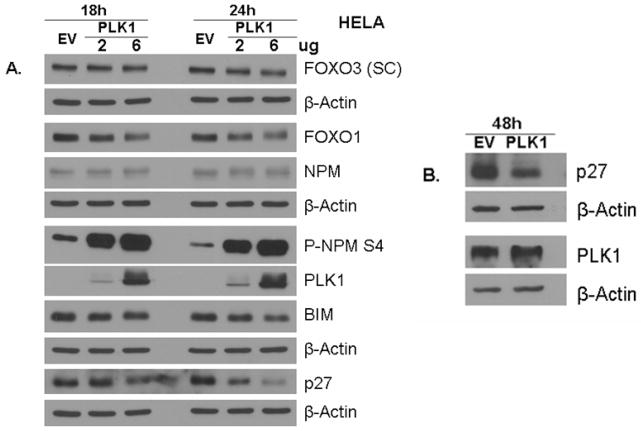
PLK1 downregulates FOXO’s targets Bim and p27
Since PLK1 induces FOXO3 exclusion from the nucleus, we further investigated its effect on FOXO3-dependent expression targets Bim and p27. We demonstrate here that PLK1 expression induces a decrease in expression of both Bim and p27 (Figure 5A & B). Moreover, PLK1 induces a decrease in total FOXO3 and FOXO1 levels. It is well established that FOXO family members are degraded into the cytoplasm by an ubiquitin-dependent mechanisms, by the proteasome1, after relocation from the nuclei to the cytoplasm. Since PLK1 induces relocalization of FOXO3 into the cytoplasm (Figure 4), this decrease in FOXO3 levels may be explained by its proteasome-dependent degradation in the cytoplasm. However, this hypothesis remains to be established.
Figure 5. PLK1 downregulates FOXO’s targets Bim and p27.
A. HeLa cells were transfected with either EV-FLAG or increasing concentrations (2μg, 6μg) of PLK1-FLAG and the expression of FOXO1, FOXO3, BIM, p27, PLK1, P-NPM1 Ser4 & NPM1 was monitored by western blot. PLK1 induces a decrease in BIM (24h) and p27 (18h and 24h), well known FOXO targets; B. HeLa cells were transfected with EV-FLAG or PLK1-FLAG. Expression of p27 was evaluated. The results show that PLK1 decreases p27 levels; β-Actin was used as a loading control; PLK1 activity is determined by measuring the phosphorylation of NPM at Ser4 (site known to be phosphorylated by PLK1)42; SC = anti-FOXO3 antibody from Santa Cruz Biotechnology;
CONCLUSIONS
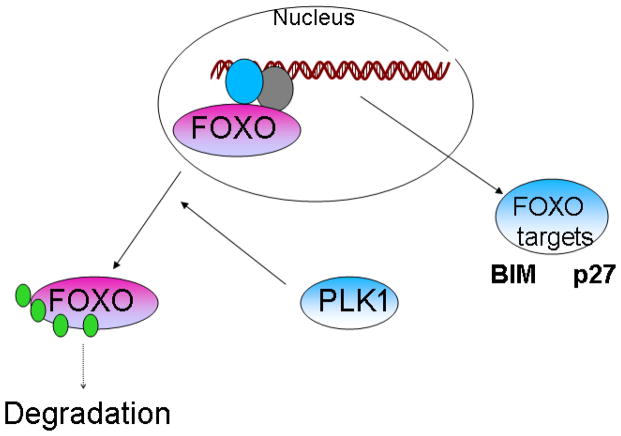
Our studies herein suggest that PLK1 forms a complex with FOXO3 and other FOXO family members and suppresses their nuclear localization and activity. The binding takes places during most phases of the cell cycle, being more prominent during the G2-M phases of the cell cycle. PLK1 is able to bind FOXO3 by using multiple binding sites, the most important region of FOXO3 being the one between the amino acids 219 and 270. PLK1 induces the exclusion of FOXO3 from the nucleus, resulting in a decrease in the expression of FOXO3-dependent targets Bim and p27, while the PLK1 knock-down can induce the opposite effect, resulting in FOXO3 accumulation into the nuclei. Moreover, PLK1 is able, at least in vitro, to phosphorylate FOXO3 (Supplementary Figure 3) at multiple sites (data not shown – Mass Spectrometry analysis of the FOXO3 bands shown in Supplementary Figure 3), suggesting that PLK1 may be a potential FOXO3 kinase. However, additional experiments are required in order to analyze the PLK1 potential to phosphorylate FOXO family members. PLK1 expression also induces a decrease in total FOXO1 and FOXO3 levels, which can be explained by our results of PLK1-dependent exclusion of FOXOs from the nucleus to the cytoplasms, where FOXO may be degraded by the proteasome1 (Figure 6).
Figure 6.
Model of how PLK1 controls FOXO3 localization, levels and activity
PLK1, considered a proto-oncogene25,47,48, is a novel promising target for the treatment of a variety of pathologies, since it is found overexpressed in many malignancies and its expression increases the risk for metastasis24–26. Several novel PLK1 inhibitors, such as BI2536, are now in clinical trials for the treatment of leukemia and other types of cancers. Our results demonstrate that PLK1 inhibits FOXO tumor suppressors, suggesting that this is a new mechanism of inactivating FOXO proteins in cancer.
Our results uncover a new inhibitory mechanism of FOXO tumor suppressor proteins in cancer. Since FOXOs are inactivated in a wide variety of malignancies, it is important to uncover druggable mechanisms of their suppression.
The TAP-MS screening identified PLK1 as a FOXO3 suppressor. Noteworthy, PLK1 inhibitors are now in clinical trials for a wide variety of malignancies.
Interestingly, it was previously suggested that PLK1 expression is controlled by FOXO3 during certain phases of cell cycle49. Our own experiments (data not shown) confirm that FOXO3 expression induces an increase in PLK1 levels. Thus, PLK1 and FOXO family members may function in a negative feedback loop, controlling both FOXO-induced cell death/cell cycle arrest and PLK1-mediated proliferation/cell cycle progression. Additional studies need to be performed in order to investigate this hypothesis.
Taken together, our results highlight the importance of a new regulatory mechanism of FOXO tumor suppressors by the PLK1 proto-oncogene.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grants CA105306, CA131664 and HL080192 to RK-F. Octavian Bucur received a fellowship from the Lady TATA Memorial Trust, London, UK. We also thank to several laboratories that provided useful reagents (see Materials and Methods section for details).
Abbreviations
- FOXOs
FOXO family members
- PLK1
Polo Like Kinase 1
- IgG
Immunoglobulin G
- Akt, PKB
Protein Kinase B
- PI3K
PhosphoInositide 3-Kinase
Footnotes
Conflict of Interest:
The authors do not declare any conflict of interest.
References
- 1.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008 Apr 7;27(16):2276–88. doi: 10.1038/onc.2008.21;. [DOI] [PubMed] [Google Scholar]
- 2.Dumitrascu GR, Bucur O. Critical physiological and pathologic al functions of Forkhead Box O tumor suppressors. Discoveries. 2013 Oct-Dec;1(1):e5. doi: 10.15190/d.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucur O, Stancu AL, Khosravi-Far R, Almasan A. Analysis of apoptosis methods recently used in Cancer Research and Cell Death & Disease publications. Cell Death Dis. 2012;3:e263. doi: 10.1038/cddis.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parody JP, Ceballos MP, Quiroga AD, Frances DE, Carnovale CE, Pisani GB, et al. FoxO3a modulation and promotion of apoptosis by interferon-α2b in rat preneoplastic liver. Liver Int. 2013 doi: 10.1111/liv.12421. [DOI] [PubMed] [Google Scholar]
- 5.Bucur O, Ray S, Bucur MC, Almasan A. APO2 ligand/tumor necrosis factor-related apoptosis-inducing ligand in prostate cancer therapy. Front Biosci. 2006 May 1;11:1549–68. doi: 10.2741/1903. [DOI] [PubMed] [Google Scholar]
- 6.Bucur O, Nat R, Cretoiu D, Popescu LM. Phagocytosis of apoptotic cells by microglia in vitro. J Cell Mol Med. 2001 Oct-Dec;5(4):438–41. doi: 10.1111/j.1582-4934.2001.tb00181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raval FM, Nikolajczyk BS. The Bidirectional Relationship between Metabolism and Immune Responses. Discoveries. 2013 Oct-Dec;1(1):e6. doi: 10.15190/d.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan JA, Kim EH, Plisch EH, Peng SL, Suresh M. FOXO3 regulates CD8 T cell memory by T cell-intrinsic mechanisms. PLoS Pathog. 2012;8(2):e1002533. doi: 10.1371/journal.ppat.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seiler F, Hellberg J, Lepper PM, Kamyschnikow A, Herr C, Bischoff M, et al. FOXO transcription factors regulate innate immune mechanisms in respiratory epithelial cells. J Immunol. 2013;190(4):1603–13. doi: 10.4049/jimmunol.1200596. [DOI] [PubMed] [Google Scholar]
- 10.Yeo H, Lyssiotis CA, Zhang Y, Ying H, Asara JM, Cantley LC, Paik JH. FoxO3 coordinates metabolic pathways to maintain redox balance in neural stem cells. EMBO J. 2013;32(19):2589–602. doi: 10.1038/emboj.2013.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calnan DR, Webb AE, White JL, Stowe TR, Goswami T, Shi X, et al. Methylation by Set9 modulates FoxO3 stability and transcriptional activity. Aging (Albany NY) 2012 Jul;4(7):462–79. doi: 10.18632/aging.100471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang JY, Hung MC. Deciphering the role of forkhead transcription factors in cancer therapy. Curr Drug Targets. 2011;12(9):1284–1290. doi: 10.2174/138945011796150299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jagani Z, Singh A, Khosravi-Far R. FoxO tumor suppressors and BCR-ABL-induced leukemia: a matter of evasion of apoptosis. Biochim Biophys Acta. 2008;1785(1):63–84. doi: 10.1016/j.bbcan.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calautti E. Akt modes of stem cell regulation: more than meets the eye? Discoveries. 2013 Oct-Dec;1(1):e8. doi: 10.15190/d.2013.8;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh A, Ye M, Bucur O, Zhu S, Tanya Santos M, et al. Protein phosphatase 2A reactivates FOXO3a through a dynamic interplay with 14-3-3 and AKT. Mol Biol Cell. 2010;21(6):1140–52. doi: 10.1091/mbc.E09-09-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villén J, Becker EB, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006 Jun 2;125(5):987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 17.Tuteja G, Kaestner KH. SnapShot: forkhead transcription factors I. Cell. 2007;130(6):1160. doi: 10.1016/j.cell.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008 Aug 5;105(31):10762–7. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008 Feb;10(2):138–48. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greer EL, Brunet A. FOXO transcription factors in ageing and cancer. Acta Physiol (Oxf) 2008 Jan;192(1):19–28. doi: 10.1111/j.1748-1716.2007.01780.x;. [DOI] [PubMed] [Google Scholar]
- 21.Skapek SX, Anderson J, Barr FG, Bridge JA, Gastier-Foster JM, Parham DM, et al. PAX-FOXO1 fusion status drives unfavorable outcome for children with rhabdomyosarcoma: a children’s oncology group report. Pediatr Blood Cancer. 2013 Sep;60(9):1411–7. doi: 10.1002/pbc.24532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jagani Z, Song K, Kutok JL, Dewar MR, Melet A, Santos T, et al. Proteasome inhibition causes regression of leukemia and abrogates BCR-ABL-induced evasion of apoptosis in part through regulation of forkhead tumor suppressors. Cancer Res. 2009 Aug 15;69(16):6546–55. doi: 10.1158/0008-5472.CAN-09-0605;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bucur O, Stancu AL, Goganau I, Petrescu SM, Pennarun B, et al. Combination of bortezomib and mitotic inhibitors down-modulate Bcr-Abl and efficiently eliminates tyrosine-kinase inhibitor sensitive and resistant Bcr-Abl-positive leukemic cells. PLoS One. 2013;8(10):e77390. doi: 10.1371/journal.pone.0077390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cholewa BD, Liu X, Ahmad N. The Role of Polo-like Kinase 1 in Carcinogenesis: Cause or Consequence? Cancer Res. 2013;73(23):6848–55. doi: 10.1158/0008-5472.CAN-13-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang G, Zhang Z, Liu Z. Polo-like kinase 1 is overexpressed in renal cancer and participates in the proliferation and invasion of renal cancer cells. Tumour Biol. 2013;34(3):1887–94. doi: 10.1007/s13277-013-0732-0. [DOI] [PubMed] [Google Scholar]
- 26.Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6(4):321–30. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- 27.Dibb M, Han N, Choudhury J, Hayes S, Valentine H, West C, Ang YS, Sharrocks AD. The FOXM1-PLK1 axis is commonly upregulated in oesophageal adenocarcinoma. Br J Cancer. 2012;107(10):1766–75. doi: 10.1038/bjc.2012.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Didier C, Demur C, Grimal F, Jullien D, Manenti S, Ducommun B. Evaluation of checkpoint kinase targeting therapy in acute myeloid leukemia with complex karyotype. Cancer Biol Ther. 2012;13(5):307–13. doi: 10.4161/cbt.19074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg T, Bug G, Ottmann OG, Strebhardt K. Polo-like kinases in AML. Expert Opin Investig Drugs. 2012;21(8):1069–74. doi: 10.1517/13543784.2012.691163. [DOI] [PubMed] [Google Scholar]
- 30.Gleixner KV, Ferenc V, Peter B, Gruze A, Meyer RA, Hadzijusufovic E, et al. Polo-like kinase 1 (Plk1) as a novel drug target in chronic myeloid leukemia: overriding imatinib resistance with the Plk1 inhibitor BI 2536. Cancer Res. 2010;70(4):1513–23. doi: 10.1158/0008-5472.CAN-09-2181. [DOI] [PubMed] [Google Scholar]
- 31.Lund-Andersen C, Patzke S, Nähse-Kumpf V, Syljuåsen RG. PLK1-inhibition can cause radiosensitization or radioresistance dependent on the treatment schedule. Radiother Oncol. 2014 Feb;110(2):355–61. doi: 10.1016/j.radonc.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 32.Bucur O, Pennarun B, Stancu AL, Nadler M, Muraru MS, Bertomeu T, Khosravi-Far R. Poor antibody validation is a challenge in biomedical research: a case study for detection of c-FLIP. Apoptosis. 2013 Oct;18(10):1154–62. doi: 10.1007/s10495-013-0880-0;. [DOI] [PubMed] [Google Scholar]
- 33.Pennarun B, Gaidos G, Bucur O, Tinari A, Rupasinghe C, Jin T, et al. killerFLIP: a novel lytic peptide specifically inducing cancer cell death. Cell Death Dis. 2013 Oct 31;4:e894. doi: 10.1038/cddis.2013.401;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruyère C, Meijer L. Targeting cyclin-dependent kinases in anti-neoplastic therapy. Curr Opin Cell Biol. 2013;25(6):772–9. doi: 10.1016/j.ceb.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2(1):81–91. doi: 10.1016/S1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- 36.Tang ED, Nuñez G, Barr FG, Guan KL. Negative regulation of the forkhead transcription factor FKHR by Akt. J Biol Chem. 1999 Jun 11;274(24):16741–6. doi: 10.1074/jbc.274.24.16741. [DOI] [PubMed] [Google Scholar]
- 37.van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279(28):28873–9. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 38.Ray S, Bucur O, Almasan A. Sensitization of prostate carcinoma cells to Apo2L/TRAIL by a Bcl-2 family protein inhibitor. Apoptosis. 2005;10(6):1411–8. doi: 10.1007/s10495-005-2490-y. [DOI] [PubMed] [Google Scholar]
- 39.SiShi L, Buchbinder E, Wu L, Bjorge JD, Fujita DJ, Zhu S. EGFR and HER2 levels are frequently elevated in colon cancer cells. Discoveries Reports. 2014 Sep-Dec;1(1):e1. doi: 10.15190/drep.2014.1. [DOI] [Google Scholar]
- 40.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8(10):1064–73. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 41.Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta. 2011;1813(11):1938–45. doi: 10.1016/j.bbamcr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Zhang H, Shi X, Paddon H, Hampong M, Dai W, Pelech S. B23/nucleophosmin serine 4 phosphorylation mediates mitotic functions of polo-like kinase 1. J Biol Chem. 2004 Aug 20;279(34):35726–34. doi: 10.1074/jbc.M403264200. [DOI] [PubMed] [Google Scholar]
- 43.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003 Feb 21;299(5610):1228–31. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 44.Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003 Oct 3;115(1):83–95. doi: 10.1016/S0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 45.Vacha P, Zuskova I, Bumba L, Herman P, Vecer J, Obsilova V, Obsil T. Detailed kinetic analysis of the interaction between the FOXO4-DNA-binding domain and DNA. Biophys Chem. 2013 Dec 31;184:68–78. doi: 10.1016/j.bpc.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Keniry M, Pires MM, Mense S, Lefebvre C, Gan B, Justiano K, et al. Survival factor NFIL3 restricts FOXO-induced gene expression in cancer. Genes Dev. 2013 Apr 15;27(8):916–27. doi: 10.1101/gad.214049.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu LY, Yu X. The balance of Polo-like kinase 1 in tumorigenesis. Cell Div. 2009 Jan 22;4:4. doi: 10.1186/1747-1028-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mok WC, Wasser S, Tan T, Lim SG. Polo-like kinase 1, a new therapeutic target in hepatocellular carcinoma. World J Gastroenterol. 2012;18(27):3527–36. doi: 10.3748/wjg.v18.i27.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alvarez B, Martínez-A C, Burgering BM, Carrera AC. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature. 2001;413(6857):744–7. doi: 10.1038/35099574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.