Abstract
The genus Acinetobacter consists of 31 validly published species ubiquitously distributed in nature and primarily associated with nosocomial infection. We report the 3.4 Mb genome of Acinetobacter haemolyticus strain MTCC 9819T. The genome has a G + C content of 40.0% and includes 3 rRNA genes (5S, 23S, 16S) and 65 aminoacyl-tRNA synthetase genes.
Keywords: Acinetobacter haemolyticus strain MTCC 9819T, Whole genome, Illumina-HiSeq 1000 technology, CLCbio wb6, Rapid annotations using subsystems technology (RAST)
| Specifications | |
|---|---|
| Organism/cell line/tissue | Acinetobacter haemolyticus |
| Strain(s) | MTCC 9819T |
| Sequencer or array type | Sequencer; the Illumina-HiSeq 1000 |
| Data format | Processed |
| Experimental factors | Microbial strain |
| Experimental features | Whole genome sequencing of A. haemolyticus strain MTCC 9819T, assembly and annotation |
| Consent | n/a |
Direct link to the data
Direct link: http://www.ncbi.nlm.nih.gov/nuccore/ASYX00000000.
Genus Acinetobacter was proposed by Brisou and Pre'vot in 1954 [1]. This genus comprises Gram-negative, strictly aerobic, nonfermenting, nonfastidious, nonmotile, catalase-positive, oxidase-negative bacteria with a DNA G + C content of 39% to 47% [2]. According to Euzeby's list of prokaryotic names with standing in nomenclature (http://www.bacterio.cict.fr/a/acinetobacter.html) the genus Acinetobacter consists of 31 validly published species. Acinetobacter haemolyticus was proposed by Bouvet and Grimont 1986 [3]; it was isolated from human clinical specimens and environment, with characteristics corresponding to those of the genus Acinetobacter. The organism in this study is Acinetobacter haemolyticus strain MTCC 9819T equivalent to DSM 6962 (= ATCC 17906, CIP 64.3, NCTC 10305).
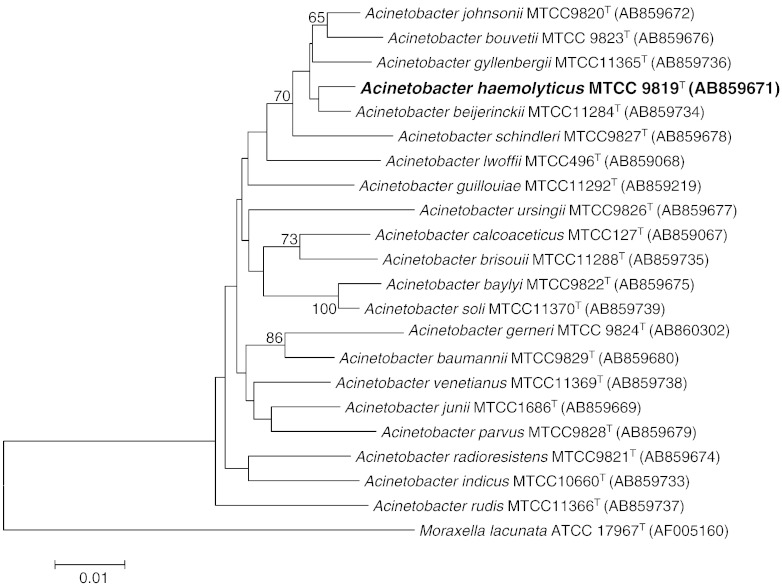
A. haemolyticus strain MTCC 9819T was obtained from MTCC and grown on tryptic soya agar medium (TSA; HiMedia) at 30 °C. Genomic DNA was extracted from 36 hour old culture using ZR Fungal/Bacterial DNA MiniPrep™ as per manufacturer's instructions. Identification was reconfirmed using 16S rRNA sequencing. Amplification and sequencing of 16S rRNA was performed as described by Mayilraj et al. 2006 [4]. To determine the phylogenetic relationship of strain MTCC 9819T, the 16S rRNA sequence consisting of 1502 bp was compared with those of type strains of species of related genera and identification of phylogenetic neighbors and the calculation of pairwise 16S rRNA gene sequence similarities were achieved using the EzTaxon server [5] and aligned using mega version 5.0 [6]. Phylogenetic trees were constructed using the neighbor-joining algorithm. Bootstrap analysis was performed to assess the confidence limits of the branching (Fig. 1.).
Fig. 1.
Phylogenetic tree constructed using the neighbor-joining algorithm, shows the position of A. haemolyticus strain MTCC 9819T relative to the type strains of the other species within the genus Acinetobacter.
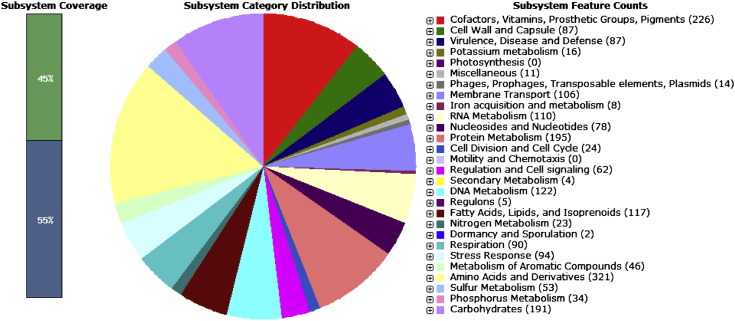
The genome of A. haemolyticus strain MTCC 9819T was sequenced using Illumina-HiSeq 1000 technology. Sequencing resulted in 5,755,416 paired-end reads (insert size of 350 bp) of length 101 bp. A total of 5,580,067 high-quality reads with approximately 166 × coverage were assembled with CLCbio wb6 (word size = 50 and bubble size = 100) and to obtain 182 contigs (N50, 57,666 bp) of 3,384,229 bp with an average GC content of 40%. The functional annotation was carried out by RAST (rapid annotation using subsystem technology) [7], Fig. 2 shows the subsystem distribution of strain A. haemolyticus strain MTCC 9819T, and tRNA was predicted by tRNAscan-SE 1.23 [8] and rRNA genes by RNAmmer 1.2 [9]. The genome contains 3 rRNA genes (5S, 23S, 16S) and 65 aminoacyl-tRNA synthetase genes. A total of 3122 coding regions (1546 transcribed from the positive strand and 1576 from the negative strand) were found in the genome, of which 2298 (74%) could be functionally annotated. The genome coding density is 87% with an average gene length of 911 bp. The annotated genome has 65 genes responsible for resistance to antibiotic and toxic compounds including 13 genes for MDR efflux pumps. One hundred and six genes code for membrane transport proteins. Forty four genes are involved in response to oxidative stress, 10 for osmotic stress response and 16 genes for heat shock and many more stress responses, all summed up to 94 genes for stress response are present.
Fig. 2.
Sub-system distribution of strain A. haemolyticus strain MTCC 9819T (based on RAST annotation server).
The functional comparison of genome sequences available on the RAST server revealed the closest neighbors of A. haemolyticus strain MTCC 9819T as Acinetobacter junii SH205 (score 503) followed by Acinetobacter baumanii ACICU (score 489), Acinetobacter haemolyticus ATCC 19194 (score 476) and Acinetobacter baumanii AB0057 (score 453).
Nucleotide sequence accession number
The A. haemolyticus strain MTCC 9819T whole genome shot gun (WGS) project has been deposited at DDBJ/EMBL/GenBank under the project accession ASYX00000000 of project (01) which has the accession numbers ASYX01000000 and consists of sequences ASYX01000001 ASYX01000182.
Conflict of interest
The authors declare that there is no conflict of interest on any work published in this paper.
Acknowledgments
This work was funded by CSIR-IMTECH. N.K.S. is supported by a University Grant Commission (UGC) fellowship, and I.K. is supported by a research fellowship from CSIR. We thank the Sandor next-generation genomics facility for the help in obtaining the genome sequence. This is IMTECH communication number 0107/2013.
References
- 1.Brisou J., Prevot A.R. Etudes de systematique bacterienne. X. Revision des speces reunies dans le genre Achromobacter. Ann. Inst. Pasteur. 1954;86:722–728. [PubMed] [Google Scholar]
- 2.Peleg A.Y., Seifert H., Paterson D.L. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouvet P.J.M., Grimont P.A.D. Taxonomy of the Genus Acinetobacterwith the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Evol. Microbiol. 1986;36(2):228–240. [Google Scholar]
- 4.Mayilraj S., Saha P., Korpole S., Saini H.S. Ornithinimicrobium kibberense sp. nov. isolated from the Himalayas, India. Int. J. Syst. Evol. Microbiol. 2006;56:1657–1661. doi: 10.1099/ijs.0.64138-0. [DOI] [PubMed] [Google Scholar]
- 5.Kim O., Cho Y.J., Lee K., Yoon S.H., Kim M., Na H., Park S.C., Jeon Y.S., Lee J.H., Yi H., Won S., Chun J. Introducing EzTaxon-e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 6.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., Meyer F., Olsen G.J., Olson R., Osterman A.L., Overbeek R.A., McNeil L.K., Paarmann D., Paczian T., Parrello B., Pusch G.D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagesen K., Hallin P., Rodland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]