Abstract
To assess the potential competitive endogenous RNA (ceRNA) network in hepatocellular cancer (HCC), the lncRNA, mRNA, and microRNA microarrays were conducted on 3 pairs of HCC and paired normal liver tissue. After that, the arrays were normalized and analyzed with gene oncology (GO) and pathway analysis. Next, we screened out the pseudogenes and their cognate protein coding genes which are both down-regulated in HCC. Finally, the up-regulated microRNA binding sites were predicted on the most down-regulated pseudogene and its cognate protein-coding gene. All the array data were uploaded to Gene Expression Omnibus (accession number GSE64633).
Keywords: lncRNA, ceRNA, Hepatocellular carcinoma
| Specifications | |
|---|---|
| Organism cell line/tissue | Homo sapiens |
| Sex | Male or female |
| Sequencer or array type | Arraystar Human LncRNA Microarray V2.0, miRCURY™ LNA Array (v.18.0) |
| Data format | Raw data: TXT. SOFT. MINiNAL. file |
| Experimental factors | Tumor vs. normal liver tissue in humans |
| Experimental features | Use whole genome microarray to predict the ceRNA network in HCC |
| Consent | All patients gave their written informed consent before study entry. |
| Sample source location | Guangzhou, China |
Direct link to deposited data
Experimental design, materials and methods
Human tissues
After informed consent was given and under the Institutional Review Board approval, human HCC and paired normal liver tissues were obtained immediately after surgery at the Johns Hopkins Hospital (Baltimore, US), The 3rd Affiliated Hospital of Sun Yat-sen University (Guangzhou, China), Fundeni Clinical Institute (Bucharest, Romania), and Ion Chiricuta Comprehensive Cancer Center (Clui Nanoca, Romania). Tissues were snap frozen upon acquisition and stored in a − 80 °C freezer until use.
LncRNA, mRNA labeling and array hybridization
The Arraystar Human lncRNA Array v2.0 was used to profile both lncRNAs and messenger RNAs (mRNAs) in human genome of 3 pairs of human HCC and the matched normal tissues. 33,045 LncRNAs were collected from the authoritative data sources including RefSeq, UCSC KnownGenes, Ensembl and many related literatures. Sample labeling and array hybridization were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology) with minor modifications. Briefly, mRNA was purified from 1 μg total RNA after the removal of rRNA (mRNA-ONLY™ Eukaryotic mRNA Isolation Kit, Epicentre). Then, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3′ bias utilizing a random priming method. The labeled cRNAs were purified by the RNAeasy Mini Kit (Qiagen). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured by using NanoDrop ND-1000. 1 μg of each labeled cRNA is fragmented by adding 11 μl 10 × Blocking Agent and 2.2 μl of 25 × Fragmentation Buffer, and then heated the mixture at 60 °C for 30 min; finally 55 μl 2 × GE Hybridization Buffer is added to dilute the labeled cRNA. 100 μl of hybridization solution was dispensed into the gasket slide and assembled to the lncRNA expression microarray slide. The slides were incubated for 17 h at 65 °C in an Agilent hybridization oven. The hybridized arrays were washed, fixed and scanned by using the Agilent DNA Microarray Scanner (part number G2505B).
MicroRNA labeling and array hybridization
MiRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon) was used according to the manufacturer's guideline for miR labeling of the same 3 pairs of HCC and matched normal liver tissues. 1.0 mg of each sample was 3′-end-labeled with Hy3™ fluorescent label using T4 RNA ligase by the following procedure: RNA in 2.0 μl of water was combined with 1.0 μl of CIP buffer and CIP (Exiqon). The mixture was incubated for 30 min at 37 °C, and was terminated by incubation for 5 min at 95 °C. Then 3.0 μl of labeling buffer, 1.5 μl of fluorescent label (Hy3™), 2.0 μl of DMSO, and 2.0 μl of labeling enzyme were added into the mixture. The labeling reaction was incubated for 1 h at 16 °C, and terminated by incubation for 15 min at 65 °C. After stopping the labeling procedure, the Hy3™-labeled samples were hybridized on the miRCURY™ LNA Array (v.18.0) (Exiqon) according to the array manual. The total 25 μl mixture from Hy3™-labeled samples with 25 μl hybridization buffer was first denatured for 2 min at 95 °C, incubated on ice for 2 min and then hybridized to the microarray for 16–20 h at 56 °C in a 12-Bay Hybridization Systems (Hybridization System–Nimblegen Systems), which provides an active mixing action and constant incubation temperature to improve hybridization uniformity and enhance signal. Following hybridization, the slides were achieved, washed several times using Wash buffer kit (Exiqon), and finally dried by centrifugation for 5 min at 400 rpm. Then the slides were scanned using the Axon GenePix 4000B microarray scanner (Axon Instruments).
Data analysis
Agilent Feature Extraction software (version 10.7.3.1) was used to analyze lncRNA and mRNA array images. Quantile normalization and subsequent data processing were performed by using the GeneSpring GX v11.5.1 software package (Agilent Technologies). After quantile normalization of the raw data, LncRNAs and mRNAs that at least 1 out of 6 samples have flags in Present or Marginal were chosen for further data analysis. Differentially expressed LncRNAs and mRNAs with statistical significance were identified through Volcano Plot filtering. Hierarchical Clustering was performed using the Agilent GeneSpring GX software (version 11.5.1). GO analysis and Pathway analysis were performed by using the standard enrichment computation method.
For microRNA array analysis, the scanned images were then imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Replicated miRNAs were averaged and miRNAs that intensities ≥ 50 in all samples were chosen for calculating normalization factor. Expressed data were normalized using the Median normalization. After normalization, significantly differentially expressed miRNAs were identified through Volcano Plot filtering. Hierarchical clustering was performed using MEV software (v4.6, TIGR).
Discussion
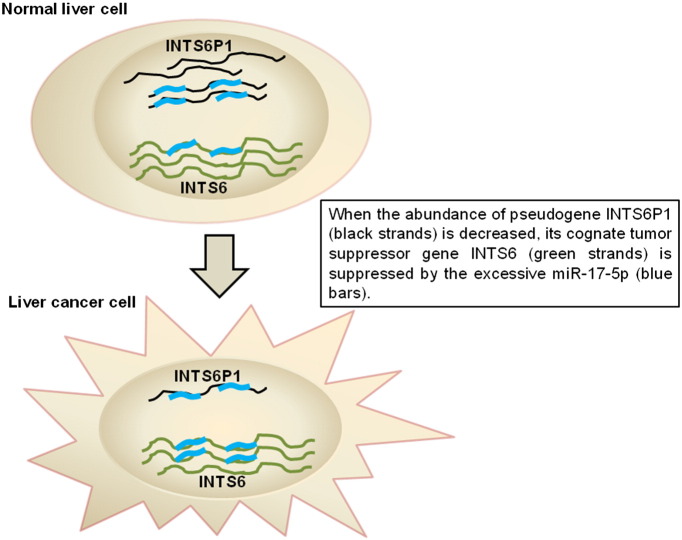
According to the ceRNA paradigm, long non-coding transcripts share miR responsive elements (MREs) with protein-coding genes, and compete with these protein-coding genes for the pool of shared miRs [1]. Thus, ceRNA would play a crucial role in the regulation of their cognate genes [2], [3], [4]. Here, we described a dataset composed of lncRNA, mRNA and microRNA microarray gene expression profiling for HCC and normal liver tissue. With this dataset, we were able to find out some ceRNA networks with tumor suppressive potential. We believe that this dataset would be particularly valuable for investigating the interplay among lncRNA, mRNA, and microRNA in HCC. For example, we have found that INTS6P1 and INTS6 are part of the same competitive tumor suppressor network that includes oncomiR-17-5p (Fig. 1). This finding provides a new insight in understanding the underlying mechanisms of hepatocarcinogenesis and uncovers the potential therapeutic targets.
Fig. 1.
Working hypothesis postulates that INTS6, INTS6P1 and miR-17-5p are part of same regulatory circuit. INTS6P1 levels (Step 1), through modulation of miR-17-5p levels (Step 2) impacts the levels of INTS6 (Step 3). Decreasing levels of INTS6P1 (red arrow) results in increasing levels of miR-17-5p (blue bar) with subsequent decreasing levels of tumor suppressor INTS6. Increasing levels of INTS6P1 (green arrow) results in decreasing levels of miR-17-5p with subsequent increasing levels of tumor suppressor INTS6.
Contributor Information
Minqiang Lu, Email: larrylmq@gmail.com.
Florin M. Selaru, Email: fselaru1@jhmi.edu.
References
- 1.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karreth F.A., Tay Y., Perna D., Ala U., Tan S.M., Rust A.G., DeNicola G., Webster K.A., Weiss D., Perez-Mancera P.A. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tay Y., Kats L., Salmena L., Weiss D., Tan S.M., Ala U., Karreth F., Poliseno L., Provero P., Di Cunto F. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang L., Du W.W., Yang X., Chen K., Ghanekar A., Levy G., Yang W., Yee A.J., Lu W.Y., Xuan J.W. Versican 3′-untranslated region (3′-UTR) functions as a ceRNA in inducing the development of hepatocellular carcinoma by regulating miRNA activity. FASEB J. 2013;27:907–919. doi: 10.1096/fj.12-220905. [DOI] [PubMed] [Google Scholar]