Abstract
Kuwaiti native population comprises three distinct genetic subgroups of Persian, “city-dwelling” Saudi Arabian tribe, and nomadic “tent-dwelling” Bedouin ancestry. Bedouin subgroup is characterized by presence of 17% African ancestry; it owes it origin to nomadic tribes of the deserts of Arabian Peninsula and North Africa. By sequencing whole genome of a Kuwaiti male from this subgroup at 41X coverage, we report 3,752,878 SNPs, 411,839 indels, and 8451 structural variations. Neighbor-joining tree, based on shared variant positions carrying disease-risk alleles between the Bedouin and other continental genomes, places Bedouin genome at the nexus of African, Asian, and European genomes in concordance with geographical location of Kuwait and Peninsula. In congruence with participant's medical history for morbid obesity and bronchial asthma, risk alleles are seen at deleterious SNPs associated with obesity and asthma. Many of the observed deleterious ‘novel’ variants lie in genes associated with autosomal recessive disorders characteristic of the region.
Keywords: Whole genome sequence, Arabian Peninsula, Nomadic Bedouin ancestry, Kuwaiti population, Intergenome distances, “Tent-dwelling” Bedouins
Introduction
Population of Kuwait comprises early settlers that include tribes from Arabian and Persian countries, and nomadic Bedouins of the desert [1]. By way of analyzing genome-wide genotypes from 273 Kuwaiti natives, we recently demonstrated three distinct genetic subgroups in Kuwaiti population [2]: Kuwait P (KWP) of Persian ancestry; Kuwait S (KWS) of “city-dwelling” Saudi Arabian tribe ancestry, and Kuwait B (KWB) that includes most of the “tent-dwelling” Bedouin participants (recruited to provide samples for genotyping). The KWB is distinguished from the other two groups by a characteristic presence of 17% African ancestry (ranging from 11.7% to 39.4%); Arabian ancestry is seen more in the Saudi Arabian tribe ancestry subgroup (at 69%) than in the Bedouin group (at 40%). Populations from other states of the Arabian Peninsula also display such a characteristic presence of African ancestry: (i) analysis of mitochondrial DNA variation in Saudi Arabian samples reveals that the Saudi Arabian population harbors as much as 20% genetic contribution from Africa [3]; (ii) analysis of Saudi Arabian Y-chromosome data indicates that around 14% of the Saudi Arabian Y-chromosome pool is typical of African biogeography ancestry [4]; (iii) analysis of mitochondrial DNA variation in populations from Near East and Africa identifies a very high frequency of African lineages (specifically sub-Saharan) in the Yemen Hadramawt [5]; and (iv) analysis of genome-wide genotypes in individuals from Qatar identifies three clear clusters of genotypes with the third cluster comprising individuals with high African admixture [6].
Bedouins are “tent-dwelling” nomads who roamed the deserts of Middle East; they epitomize the best adaptation of human life to desert conditions [7]. In much of the Middle East and North Africa, the term Bedouin is used to descriptively differentiate between those (bedu) whose livelihood is based on raising livestock by mainly natural graze and those (hadar) who have an agricultural or urban base [8]. Bedouins are originally desert-dwelling tribes of the Arabian Peninsula and are particularly descendants of (i) those settled in the southwestern Arabia, in the mountains of Yemen; and (ii) those settled in North-Central Arabia. Bedouins started to spread out to surrounding deserts of Middle East (particularly Arabian and Syrian deserts) and North Africa (particularly Sinai Peninsula of Egypt and the Sahara Desert of North Africa) due to repeated droughts, growing population and tribal wars. While the “pure” urban-dwelling Arabian tribes formed the leadership class and owned vast amounts of lands, the nomadic Bedouins often worked in the lands of the Arab tribes or tended sheep and camels and moved from one location to another in search of grazing grounds. The Bedouins, as tradition dictated, often married cousins. Marrying within the family helped strengthen bonds among extended families struggling to survive the desert. This centuries-old custom of intermarriage has had devastating genetic effects [9].
The Kuwait Genome Project (KGP) aims to sequence genomes from the three different ethnic subgroups inhabiting Kuwait. In this paper, we report, for the first time, genome sequence resource from the Bedouin subgroup by sequencing a whole genome at 40.96 × coverage. The participant that provided the sample is of Yemeni Bedouin ancestry from Kuwait. We catalog 3,752,878 SNPs, 411,839 short indels and 8451 structural variations. We further present neighbor-joining trees that depict intergenome comparisons between the genomes of nomadic Bedouins, “city-dwelling” Arabian tribes, and other continental populations.
Results
We sequenced whole genome of a 20 year old male (of Yemeni origin) from the Kuwaiti Bedouin subgroup using Illumina HiSeq 2000. We generated 1273.08 million paired-end reads of length 101 bps that were aligned to the human reference genome hg19. 95.57% of the reads were mapped to the reference genome, resulting in coverage of 41 × (Supplementary Table S1).
Ancestry estimation and haplogroup analysis
Examination of genetic clusters derived using principal component analysis (PCA) for Kuwait population (Fig. 1) reveals that this sample is located deep in the Bedouin cluster (and not at boundaries of the clusters or in regions that overlap among the three clusters). The surname lineage classification identified the participant as a desert-dwelling Bedouin tribe. Further, the ancestry composition of the genetic makeup of the KWB individual is seen as: European (French_Basque)—11%; Arab (Negev Bedouin)—44.7%; sub-Saharan African (Biaka_Pygmies)—17.3%; and West Asia (Druze, Brahui)—24.8%. This is consistent with the observed compositions in the Bedouin substructure of Kuwaiti population [2]: European (French_Basque)—11%; Arab (Negev Bedouin)—45.0%; sub-Saharan African (Biaka_Pygmies)—17.0%; and West Asia (Druze, Brahui)—25%. As stated in the Introduction, the nomadic Bedouin subgroup (KWB) is distinguished from the other two groups by a characteristic presence of 17% African ancestry. Arabian ancestry is seen more in the Saudi Arabian tribe ancestry subgroup (KWS) (at 69%) than in the Bedouin group (at 40%); and West Asian ancestry is seen more in the Persian subgroup (KWP) (at 56%) than in any of the other two groups. In order to further illustrate that the ancestry admixture composition of the Bedouin individual sequenced in this study is typical of the KWB group, we present ancestry compositions of 15 samples from the KWS subgroup [10] and one sample from the KWP subgroup [11] (Supplementary Table S2). While the sequenced Bedouin sample shows presence of 17.3% African ancestry, the 16 samples from the other two subgroups show African ancestry to the extent of only 0.1% to 5.1%; while the sequenced Bedouin sample shows Arabian ancestry at 44.7%; the 15 samples from the KWS subgroup shows Arabian ancestry to a large extent of 66.1% to 86.5%; and while the sequenced Bedouin sample shows only 24.8% West Asian ancestry, the sample from the KWP subgroup shows as high as 64.5%.
Fig. 1.
Scatter plot representing the first two principal components of merged data sets of the three Kuwaiti subgroups. The nomadic Bedouin sample considered for whole genome sequencing in this study is color-coded.
The KWB sample is observed to have J1e [J-P58] Y-chromosome haplogroup which is seen in the Arabian Peninsula. The overall estimated time of expansion of J1e haplogroup is around 10,000 years and the ancestors of J1e haplogroups are observed in the Caucasus and eastern Anatolian populations [12]. The frequencies of J1e in populations from the region are [12]: Sudan: (74.2%; n = 35); Yemen (67.7%; n = 62); Negev Bedouin (64.4%; n = 28); Ismaili Damascus (58.8%; n = 51); Qatar (56.9%; n = 72); Jordan (48.7%; n = 76); Sunni Hama (44.4%; n = 36); Oman (37.2%; n = 121); and UAE (34.8%; n = 164). The observed high value of 67.7% for the frequency of J1e in Yemeni population is consistent with the self-reported Yemeni ancestry by the participant.
The mitochondrial haplogroup (indication of maternal ancestry) of the Bedouin participant is determined as L3d1a1a [L3d], that is predominantly seen in West-Central Africa—among the Fulani [13], Chadians [13], Ethiopians [14], Akan people [15], Mozambique [14], and Yemen [14]. Kivisild et al. [14] analyzed mitochondrial DNA variations in 115 volunteer Yemeni donors in Kuwait (who claimed that their maternal origin was in Yemen) and found that the L macro-haplogroup (the most ancestral mitochondrial lineage) is seen in 47% of the 115 Yemeni individuals; they further found that 20 (17.4%) of the 115 Yemeni participants has the L3 mitochondrial haplogroup (that are most frequently found in sub-Saharan Africa); of these 20 participants, 6 (5.21% of 115 participants) displayed the L3d1 subclade that we observe for the individual sequenced in this study. Thus, the observation of L3d1 haplogroup for the participant in our study is consistent with Yemeni maternal origin. In order to further illustrate that the above observed L3d1 mitochondrial haplogroup is characteristic of the Bedouin sample sequenced in this study, we examined the mitochondrial haplogroups that we identified for a control group of 16 individuals from the other two subgroups of Kuwaiti population (see Supplementary Table S2); none of these 16 samples exhibit the clades of the L macro-haplogroup. Kivisild et al. [14] further compared haplotype diversity seen in Yemeni participants with those reported for Ethiopian population (East Africa); their results highlight the complexity of Ethiopian and Yemeni genetic heritage and are consistent with the introduction of maternal lineages into the South Arabian gene pool from different source populations of East Africa. Horn of Africa (a peninsula in the eastern region of the African sub-continent, enclosing Ethiopia, Somalia, Djibouti and Eritrea) is separated from the south Arabian Peninsula (particularly Yemen) by a short distance of only ~ 10 miles at the strait of Bab-el-Mandeb (the Gate of Tears); the distance across is only ~ 20 miles from Ras Menheli in Yemen to Ras Siyyan in Djibouti. Outside of Africa, L3d is mainly found in African Americans; approximately 6% of all African Americans are descendents of the L3d family line [16].
Identification of SNPs and indels
We compared the Bedouin genome with the reference human genome (hg19) for the identification of variants (SNPs and indels). We identified 4,164,717 variants, of which 3,752,878 are SNPs and 411,839 indels. We characterized the variants as ‘known’ and ‘novel’ (see Materials and methods) based on dbSNP 138 [17] annotation (which includes variants reported in 1000 Genomes Project phase I release). We find 1.94% (72,881 of 3,752,878) of the SNPs and 7.94% (32,686 of 411,839) of the indels as “novel”.
Transition-to-transversion ratio
The genome-wide transition-to-transversion (Ti:Tv) ratio, that is often used to measure specificity for SNP discovery, is 2.11 (in the case of known SNPs) and 1.98 (in the case of novel SNPs). These values are consistent with those reported in literature for whole genomes in 1000 Genomes Project and in other studies [18]—these studies observe 2–2.1 for known SNPs and 1.90–2.1 for novel SNPs.
Validation of SNP calls
We confirmed the validity of the SNP calls by utilizing the genotype data from the same sample derived using the Illumina HumanOmniExpress BeadChip (Illumina Inc, USA). The discordance in SNP calls is seen in a small number of cases (392 out of 312,694) leading to a concordance rate of SNP calls between deep sequencing experiments and genome-wide genotyping at 99.87%. The observed concordance rate is in agreement with that reported in literature—Kenna et al. [19] report a genotype concordance rate of 98.9% upon comparing genotypes for 85 variants inferred across 567 samples using Illumina highthroughput sequencing platforms with genotypes ascertained using Illumina BeadChips. Upon defining the SNPs as homozygous or heterozygous based on BeadChip calls, we find that the disagreements in the SNP calls are more often with homozygous SNPs (206 out of 392) than with heterozygous SNPs (186 out of 392) (Supplementary Table S3). As is the practice [20], we choose not to remove the inconsistent calls.
Classification of SNPs and indels based on genome annotation
Based on the locations of the variants relative to annotated genes in the genome, we classified the variants into broad classes such as intergenic, intronic, and coding variants (Supplementary Table S4). Most of the variants lie in intergenic regions (59 % of the total variants), followed by those that lie in introns, 3′ UTR and coding regions. The number of intronic SNPs (1,441,241) is around 24 times the number of exonic SNPs (61,643 comprising coding, non-coding exonic, and UTR SNPs); this is consistent with the notion that 1.1 to 1.5% of the human genome codes for exons while 26 to 30% codes for introns. We observe that SNPs from UTRs (30,043) outnumber those from coding exons of transcripts (21,616); this is consistent with observations made in studies using Illumina technology for whole-genome sequencing—e.g. Wong et al. [21] find that 0.86% of the SNPs identified through whole-genome sequencing of 100 southeast Asian Malays lie in coding exons while 1.06% lie in UTRs. Further we classified the variants from coding regions based on their effect on the encoded protein sequences (Supplementary Table S5). We identified 58 Stopgain and 12 Stoploss variants among known coding SNPs and 3 Stopgain variants among novel coding SNPs; we further identified 5 Stopgain variants among known indels and one Stopgain variant among novel indels; such Stopgain and Stoploss variants can truncate or elongate the coded peptide sequence. The 70 known SNPs that bring about loss or gain of stop codons are mapped to 74 genes. Of these 74 genes, 29 are found to be annotated in OMIM database and are associated with diseases such as Cohen syndrome, Schizophrenia, and Sepsis (Supplementary Table S6); 2 out of 3 Stopgain variants among novel SNPs, 2 out of 5 Stopgain variants from known indels and one Stopgain variant from novel indels are annotated in OMIM. 73 coding SNPs and 52 coding indels are found to disrupt splicing. We also observe that 80 of the known indels and 16 of the novel indels bring about frameshift changes in the encoded proteins.
We examined the observed 9893 nonsynonymous variants from the list of coding SNPs, using SIFT [22] and PolyPhen2 [23], to identify “potentially deleterious” variants (see Materials and methods). In this manner, we identified 2166 known and 105 novel potentially deleterious SNPs from 1841 genes. On checking functional categorization of these genes using Gene Ontology [24], we observed that the significant GO terms all point to sensory perception (such as that of olfaction and cognition) processes, neurological system process, and GPCR signaling pathways & plasma membrane (Supplementary Table S7).
In order to identify which of these potentially deleterious SNPs have been previously associated with (or implicated as causal variants for) diseases and phenotype traits, we examined (i) the NHRI GWAS Catalog, a curated resource of SNP-trait association [25], and (ii) the OMIM, a curated catalog of human genes and genetic disorders and traits with particular emphasis on molecular relationship between genetic variation and phenotypic expression [26]. A set of 48 deleterious SNPs are seen annotated for association with diseases in OMIM database and/or in GWAS Catalog (Table 1); the risk alleles at these SNPs are derived using GWAS Catalog or ClinVar [27]. Of these 48 deleterious variants associated with diseases, particularly interesting are those that are in conformity with the phenotype characteristics of the KWB participant, as detailed below:
-
(a)
Morbid obesity (at BMI of 45.5 kg/m2): The deleterious variants rs2043112 (RICTOR) [28], rs2275848 (NINJ1) [29], rs11042023 (RPL27A) [30] are associated with obesity and related traits. All these three variants carry the risk allele.
-
(b)
Abnormal waist circumference (134 cm): The deleterious variant rs1919128 (C2orf16) is associated with the bivariate trait of waist circumference—triglycerides (WC-TG) [31], and rs1545 (MKKS) is associated with the metabolic syndrome of abdominal obesity [32].
-
(c)
Bronchial asthma: The deleterious variant rs1051931 (PLA2G7) is associated with susceptibility to asthma [33]. The participant carries homozygous risk allele for the trait.
-
(d)
Family history of retinopathy: The deleterious variant rs267738 (CERS2) is associated with rhegmatogenous retinal detachment [34], and rs10151259 (RPGRIP1) is associated with Cone-rod dystrophy 13 [35]. The participant carries risk allele at the second marker. Presence of these two markers could be indicative of genetic factor for retinopathy seen in the patient's family history.
-
(e)
Smoking: The deleterious variant rs1801272 (CYP2A6) associated with smoking behavior [36].
-
(f)
Prehypertensive (at SBP/DBP of 134/73 Hg/mm2) and family history of high cholesterol: The following deleterious variants are associated with metabolic syndromes: rs676210 (APOB) associated with LDL [37], rs6756629 (ABCG5) associated with LDL [38], and rs11820589 associated with bivariate traits of TG and HDL [31]—the participant carries the risk allele at the second and third variants).
Table 1.
Deleterious SNPs annotated for association with diseases in OMIM database and/or in GWAS Catalog.
| SNPs | Geno-type | Strongest SNP-risk allele (GWAS Catalog, ClinVar, OMIM) | Mapped gene (OMIM ID) | Disease/trait; (phenotype MIM #); [inheritance]c |
|---|---|---|---|---|
| OMIM annotated variants | ||||
| rs2297950 [1:g:203194186C > T][Gly102Ser] |
het | Tb | CHIT1(600031) | Chitotriosidase deficiency; (#614122 ); [?] |
| rs1056827 [2:g:38302177C > A][Ala119Ser] a |
hom | ? | CYP1B1(601771) | Glaucoma 3A, primary open angle, congenital, juvenile, or adult onset; (#231300,#604229); [AR] |
| rs34231037 [4:g:55972946T > C][Cys482Arg ] |
het | Gb | KDR(191306) | Hemangioma, capillary infantile, susceptibility to; (#602089 ); [AD] |
| rs1573496 [4:g:100349669C > G][Gly92Ala ] |
het | ? | ADH7(103720) | Aerodigestive tract cancer, squamous cell, alcohol-related, protection against; (#103780); [MF] |
| rs1801394 [5:g:7870973A > G][Ile22Met] |
het | Gb | MTRR(602568) | Neural tube defects, folate-sensitive, susceptibility todown syndrome, susceptibility to, included; (#601634); [AR] |
| rs351855 [5:g:176520243G > A][Gly388Arg] |
hom | Ab | FGFR4(134935) | Cancer progression and tumor cell motility; (no OMIM Id); [?] |
| rs3807153 [7:g:138417791A > G][Met580Thr] |
het | Gb | ATP6V0A4(605239) | Renal tubular acidosis, distal; (#602722); [AR] |
| rs1801968 [9:g:132580901C > G][Asp216His] |
het | Gb | TOR1A(605204) | Dystonia-1, modifier of Dystonia-1, torsion; (#128100); [AD] |
| rs1800450 [10:g:54531235C > T][Gly54Asp] |
het | Tb | MBL2(154545) | Chronic infections, due to MBL deficiency; (#614372); [?] |
| rs3135506 [11:g:116662407G > C][Ser19Trp] |
het | Cb | APOA5(606368) | Hypertriglyceridemia, susceptibility to; (#145750); [AD} |
| rs7308720 [12:g: 40657700C > G][Asn551Lys] |
het | Gb | LRRK2(609007) | Parkinson disease 8; (#607060); [AD] |
| rs2232387 [12:g:52827608C > T][Ala12Thr] |
het | Tb | KRT75(609025) | Pseudofolliculitis barbae, susceptibility to; (#612318); [?] |
| rs10151259 [14:g:21790040G > T][Ala547Ser] a |
het | Tb | RPGRIP1(605446) | Cone-rod dystrophy 13; (#608194); [AR] |
| rs3743930 [16:g:3304626C > G][Glu148Gln] |
het | Gb | MEFV(608107) | Familial Mediterranean fever, AD, familial Mediterranean fever, AR; (#134610,#249100); [AD; AR] |
| rs4673 [16:g:88713236A > G][Tyr72His] |
het | Gb | CYBA(608508) | Chronic granulomatous disease, (#233690); [AR] |
| rs6504649 [17:g:48437456C > G][Thr801Arg] |
het | Gb | XYLT2(608125) | Pseudoxanthoma elasticum, modifier of severity of; (#264800); [AR] |
| rs1545 [20:g:10386013C > A][Gly532Val] a |
het | ? | MKKS(605552) | Abdominal obesity—metabolic syndrome; (%605552); [?] |
| rs1801265 [1:g:98348885G > A][Arg29Cys] |
hom | Ab | DPYD(612779) | Dihydropyrimidine dehydrogenase deficiency; (#274270); [AR] |
| rs486907 [1:g:182554557C > T][Arg462Gln] |
hom | Tb | RNASEL(180435) | Prostate cancer 1; (#601518); [AD] |
| rs2286963 [2:g:211060050T > G][Lys333Gln] |
het | Gb | ACADL | Metabolite levels; [?] |
| rs6180 [5:g:42719239A > C][Ile526Leu] a |
het | Cb | GHR(600946) | Hypercholesterolemia, familial, modification of; (#143890); [AD] |
| rs1051931 [6:g:46672943A > G][Val379Ala] a |
hom | Gb | PLA2G7(601690) | Asthma, susceptibility to, Atopy;(#600807, #147050); [AD, MF] |
| rs7133914 [12:g:40702911G > A][Arg1398His] |
het | Ab | LRRK2(609007) | Parkinson disease 8; (#607060); [AD] |
| rs10246939 [7:g:141672604T > C][Ile296Val] |
hom | Cb | TAS2R38(607751) | Phenylthiocarbamide tasting; (#171200); [AD] |
| rs61751507 [10:g: 101829514C > T][Gly178Asp] |
Het | Tb | CPN1(603103) | Carboxypeptidase N deficiency; (#212070); [AR] |
| rs3827103 [20:g:54824029G > A][Val81Ile] |
Het | ? | MC3R(155540) | Mycobacterium tuberculosis, protection against; (%612929); [?] |
| GWAS annotated variants | ||||
| rs3811444 [1:g:248039451C > T][Thr374Met] |
hom | Ab | CERS2 | Platelet counts, red blood cell traits; [?] |
| rs676210 [2:g:21231524G > A][Pro2739Leu] a |
het | G | APOB | LDL (oxidized), lipid metabolism phenotypes; [?] |
| rs6756629 [2:g:44065090G > A][Arg50Cys] a |
het | G | ABCG5 | Cholesterol total, LDL cholesterol (protective effect?); [?] |
| rs2043112 [5:g:38955796G > A][Ser837Phe] a |
het | Ab | RICTOR | Obesity-related traits; [?] |
| rs240768 [6:g:100957344T > C][Tyr2176Cys] |
het | T | ASCC3 | Economic and political preferences (immigration/crime); [?] |
| rs11042023 [11:g:8662516T > C][His324Arg] a |
hom | Cb | RPL27A | Obesity; [?] |
| rs11820589 [11:g:116633862G > A][Pro148Leu] a |
het | Ab | BUD13 | Metabolic syndrome (bivariate traits); [?] |
| rs3213764 [12:g:14587301A > G][Lys530Arg] |
het | Gb | ATF7IP | Prostate-specific antigen levels; [?] |
| rs2297067 [14:g:103566785C > T][Arg77Trp] |
het | Tb | EXOC3L4 | Platelet counts; [?] |
| rs2303759 [19:g:49869051T > G][Met34Arg] |
het | Cb | DKKL1 | Multiple sclerosis; [?] |
| rs267738 [1:g:150940625T > G][Glu115Ala] a |
het | A | CERS2 | Rhegmatogenous retinal detachment; [?] |
| rs1919128 [2:g:27801759A > G][Ile774Val] a |
het | A | C2orf16 | Waist circumference—triglycerides (WC-TG); [?] |
| rs2275848 [9:g:95887320G > T][Ala110Asp] a |
hom | Tb | NINJ1 | Obesity (early onset extreme); [?] |
| rs874628 [19:g:18304700A > G][Met72Val ] |
het | A | MPV17L2 | Multiple sclerosis; [?] |
| rs2239785 [22:g:36661330G > A][Glu166Lys] |
het | G | APOL1 | Glomerulosclerosis; [?] |
| Variants annotated in both OMIM and GWAS | ||||
| rs11887534 [2:g:44066247G > C][Asp19His] |
het | Cb | ABCG89(605460) | Gallstones, gallbladder disease 4; (#611465); [?] |
| rs2227564 [10:g: 75673101T > C][Leu141Pro] |
hom | Cb | ABCG8(191840, 605526) | Inflammatory bowel disease; Alzheimer disease, late-onset, susceptibility to;(#104300); [AD] |
| rs1799853 [10:g:96702047C > T][Arg144Cys] |
het | ? | CYP2C9(601130) | Warfarin maintenance dose, warfarin sensitivity; (#122700); [AD] |
| rs4149056 [12:g:21331549T > C][Val174Ala] |
het | T,C | SLCO1B1(604843) | Sex hormone-binding globulin levels, response to statin therapy; rotor type hyperbilirubinemia,; (#601816, #237450); [DR] |
| rs1801272 [19:g:41354533A > T][Leu160His] a |
het | Tb | CYP2A6(122720) | Smoking behavior, coumarin resistance (#122700); [AD] |
| rs1799990 [20:g:4680251A > G][Met129Val] |
het | A | PRNP(176640) | Prion diseases, Creutzfeldt–Jakob disease (#606688); [AD] |
| rs738409 [22:g:44324727C > G][Ile148Met] |
het | Gb | PNPLA3(609567) | Nonalcoholic fatty liver disease; (%613282); [MF] |
Abbreviations:
Variants for which the associated phenotype traits are seen with the participant (or his family) that provided sample for genome sequencing.
The alternate allele seen in the Bedouin genome corresponds to the risk allele.
AD: Autosomal dominant; AR: Autosomal recessive; MF: Multi-factorial; DR: digenic recessive;?: not known or multi-factorial.
Each of the above discussed phenotypes is seen with the Bedouin participant and the sequenced genome contains the risk alleles at one or more SNPs that are associated with each of the phenotypes. Though these genotype–phenotype associations have been demonstrated in literature and annotated in OMIM database and/or GWAS Catalog, it is imperative to mention that these individual genotype variants alone are not necessarily sufficient to account for the disorders with the Bedouin participant (for reasons mentioned below):
-
(i)
Each of the discussed phenotypes is influenced by multiple loci and multiple genetic factors, and it is often the case that a component gene can have multiple genetic variants associated with the disorder. For example, the cone-rod dystrophy (CRD) is associated with several genes (including the RPGRIP1 discussed in this work) [35]—the autosomal dominant form of CRD is associated with mutations in the peripherin/RDS, CRX, and RetGC-I genes, and the autosomal recessive form is associated with mutations in the ABCR gene. It is possible that a single locus has only a modest effect on the disease susceptibility and few or all of the reported loci may collectively participate to account for the disorder. Disease might occur only if a particular combination (pattern) of genotypes is present at different susceptibility loci [39].
-
(ii)
Data on genotype-phenotype associations, discussed in this work, come mostly from GWAS studies (Supplementary Table S8). Though GWAS studies lead to identification of associated loci, the variants that are identified need not necessarily be the ‘causal’ variants [40].
-
(iii)
The reported associations in the databases for these disorders are not necessarily demonstrated in the population of Arabian Peninsula and are very often demonstrated in European populations (see Supplementary Table S8). Such associations may not necessarily hold in ethnic populations (that are under-represented in the global genome-wide surveys) as some genetic variation is private to populations with particular continental ancestry.
We examined the genotypes at SNPs, identified as associated with the phenotypes of the Bedouin sample, in the genomes/exomes of 16 participants from the other two subgroups of Saudi Arabian tribe ancestry and Persian ancestry (see Supplementary Table S8). It is seen that for each of the studied phenotypes, the risk allele is seen in at least one another individual (from the control group) having the same phenotype. As an example: in the case of cone-rod dystrophy, 8 (out of 16) participants from the Persian and Saudi Arabian tribe ancestry subgroups have the phenotype; and 4 of these 8 patients have the alternate allele at rs10151259 (RPGRIP1) as seen in the Bedouin sample; the remaining four patients show reference allele (so are the remaining 8 participants that form the control group of unaffected participants). In these 4 patients, other mutations associated with cone-rod dystrophy might be present—efforts to identify such other mutations are out of scope for this study. We further observe that except in the cases of cone-rod dystrophy and TG-HDL phenotypes, at least one unaffected individual exhibit the risk allele—this is in concordance with the concerns listed above, particularly the concern that disease might occur only if a particular combination (pattern) of genotypes is present at different susceptibility loci [39].
Further, 9 of the annotated disorders associated with the 48 potentially deleterious SNPs are autosomal recessive. Of the 48 deleterious variants, 10 occur in homozygous form (see Table 1); the participant carries homozygous causal variants for the following two recessive disorders—(rs1056827, Glaucoma 3A [41]) and (rs1801265, dihydropyrimidine dehydrogenase deficiency [42]). Due to the practice of consanguineous marriages and inbreeding, autosomal recessive disorders are prevalent in the region.
Upon considering the 239 genes that harbor the 218 novel nonsynonymous variants, we find that 73 genes are annotated in OMIM database (Supplementary Table S9). The annotated diseases mostly include rare genetic disorders such as Myasthenia, limb-girdle, familial—autosomal recessive and congenital; Charcot–Marie–Tooth disease; Hermansky–Pudlak syndrome 3—autosomal recessive; microcephaly—autosomal recessive; mental retardation—autosomal dominant; spondyloepiphyseal dysplasia—Kimberley type; brittle cornea syndrome—autosomal recessive; deafness—autosomal recessive and congenital; Watson syndrome; congenital cataracts; nephrosis-1—congenital and Finnish type; and Mucolipidosis III gamma.
Upon examining the 9,549 noncoding SNPs for annotation in NHGRI GWAS Catalog, we find phenotype association for 26 variants (Supplementary Table S10). Many of these 26 variants are associated with phenotypes relating to diabetes, obesity and metabolite levels.
Annotation of the genome for structural variants
We identify 8451 variations consisting of 2893 deletions, 2472 duplications, 1580 insertions, 114 inversions, 470 intrachromosomal translocations, and 922 interchromosomal translocations (Supplementary Table S11). Of the total 8451 structural variations, 7672 (90.78%) are “known” structural variations, annotated in DGV (Database of Genomic Variations, a curated catalog of human genomic structural variations) [43]. Further, we see that 6696 (79.23%) of the total deletion variants lie in repeat-rich regions containing SINE (which include ALU), LINE and LTR repeat elements.
Comparison with other individual genomes
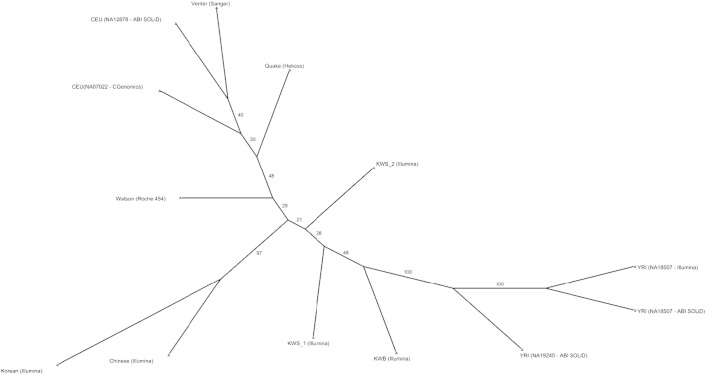
In order to assess the extent of variability that the genome of Kuwaiti subgroup of tent-dwelling Bedouin ancestry exhibits with genomes of other populations, we compare the KWB genome with two representative genomes of Kuwaiti subgroup of Saudi Arabian tribe ancestry (KWS) [10] and ten representative genomes (see Materials and methods) from four continents namely Africa (3 genomes), America (3 whites), Europe (2 whites) and Asia (1 Chinese and 1 Korean). As these 13 genomes have been sequenced using six different technologies (see Materials and methods) that have different genome coverage, the genomes cannot be directly compared to evaluate the extent of shared variants. In order to evaluate the intergenome distances among these 13 genomes, we adopt the method of Moore et al. [44] that takes care of variability across the platforms by calculating the extent of shared variant locations chromosome-by-chromosome. The consensus neighbor-joining tree derived by using this method for the 13 genomes is presented in Fig. 2. The three African sequences are closely neighbored, so are the two Asians, and the five Europeans. The two KWS genomes are clustered together and are separated from the KWB genome; all these three Kuwaiti genomes are placed amidst the five Europeans. We then examined the intersection of each of the (KWB, two Kuwaiti genomes of “city-dwelling” Saudi Arabian tribe ancestry, five Europeans, two Asians and three Africans) genome's variants with known disease-causing/predisposing alleles as cataloged in OMIM. The neighbor-joining tree based on the number of shared variant positions carrying the OMIM disease alleles is presented in Fig. 3. The OMIM variant-based tree depicts the European genomes next to one another, the Asian genomes next to one another, and the African genomes next to one another; and more importantly, the two KWS samples and the KWB genomes are near neighbors to one another and are now placed between clusters of African, and clusters of Asian (and European) genomes in congruence with the geographical location of Kuwait and the Peninsula. Of the three genomes (KWS1, KWS2, and KWB) from Kuwait, the Bedouin genome is placed closer to the African cluster in agreement with the earlier observation that the KWB is distinguished from the KWS group by a characteristic presence of 17% African ancestry.
Fig. 2.
Neighbor-joining tree based on intergenome distances calculated using genome-wide variant positions shared between the KWB genome, KWS genomes, and representative genomes from intercontinental populations.
Fig. 3.
Neighbor-joining tree based on intergenome distances calculated using variant positions associated with OMIM disease genes and shared between the KWB genome, KWS genomes, and representative genomes from intercontinental populations.
In both the trees (depicting shared genome-wide variants and shared OMIM variants), the two KWS genomes (KWS1 and KWS2) and the KWB genome are near-neighbors to one another; as these three genomes are sequenced using the same Illumina technology, it should be possible to perform direct comparisons on extent of shared variants between these two subgroups. We compared the SNPs from KWS1, KWS2 and KWB genomes; all the three genomes share a high percent of common variants with one another—KWS1 and KWB share 44.7% common variants; KWS2 and KWB share 43.1% common variants; and KWS1 and KWS2 share 45.5% common variants.
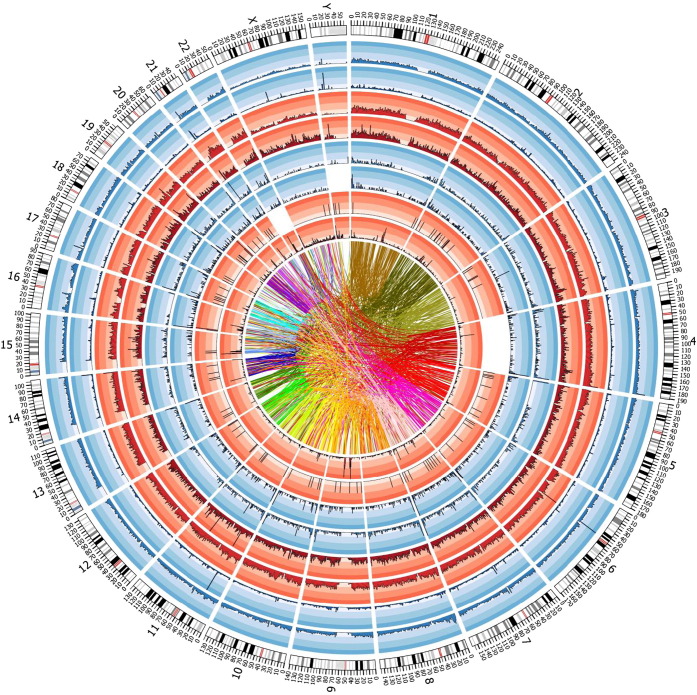
Genome view of the variants
Fig. 4 provides a high-level view of the contents of the draft genome sequence for KWB subgroup in terms of density of known and novel variants (SNPs, short and long indels) as observed from the whole genome sequence, density of duplications and the extent of chromosomal translocations. We have also created a genome browser (see the section on Data availability). The browser lets the users to view an annotated display of the identified variants and structural variations in the context of sequence and annotation tracks from other genome resources.
Fig. 4.
Summary of analysis of genomes from Kuwait subgroup of Bedouin ancestry. Tracks (from outer to inner): karyotype of human genome; density (in every window of 1 Mb) of ‘known’ SNPs (i.e. annotated in dbSNP 138); density of ‘novel’ SNPs (i.e. not annotated in dbSNP138); Density of ‘known’ indels; density of ‘novel’ indels; density of long deletions; density of long insertions; density of inversions; density of duplications; links representing intra- and interchromosomal translocations.
Discussion
Bedouins are the nomadic Arabs of the desert who live on the fringes of the Arabian Peninsula which includes parts of Kuwait, Saudi Arabia, Qatar, United Arab Emirates, Oman, Iraq, Jordan and Syria as well as Negev and Sinai desert [45]. Our earlier work [2] with genome-wide genotype data from Kuwaiti participants has shown that the Kuwaiti population is composed of three distinct genetic clusters—the first group (KWP) is largely of West Asian ancestry, representing Persians with European admixture; the second group (KWS) is predominantly of city-dwelling Saudi Arabian tribe ancestry, and the third group (KWB) includes most of the tent-dwelling Bedouin participants (recruited to provide samples for genotyping) and is characterized by the presence of 17% African ancestry; Arabian ancestry is seen more in the Saudi Arabian tribe ancestry subgroup (at 69%) than in the Bedouin group (at 40%). In this study, we consider an individual of Yemeni ancestry settled in Kuwait; the ancestry composition of the genetic makeup and the surname lineage classification of the individual are typical of the Kuwait B group. The principal component analysis places the individual near the centroid of the Kuwaiti B group. Both the Y-chromosome and the mitochondrial haplogroups are consistent with Yemeni ancestry; the observed mitochondrial haplogroup of L3d1a1a [L3d] is predominantly seen in West-Central Africa; and the J1e [J-P58] Y-chromosome haplogroup is seen in high frequencies in states from the Arabian Peninsula.
The whole genome of the Bedouin individual is sequenced at a high-depth coverage of > 40 ×. Validation of identified SNPs led to a concordance rate of 98.9% between sequencing and BeadChip array genotyping results. Up to 96% of the identified SNPs and indels are validated in the dbSNP 138 database. We believe the remaining 72,881 novel SNPs and 32,686 novel indels add to the repertoire of observed human genome variations. Further, of the identified 8451 structural variations, 779 are novel (i.e. not annotated in DGV, Database of Genomic Variations). We believe that functional level analysis of such population-dependent genomic variations may further shed light on disease mechanisms. Neighbor-joining tree constructed using intergenome distances, calculated based on shared disease-causing variants, between the Bedouin genome and continental genomes places the Bedouin genome (along with the two Kuwaiti genomes of Saudi Arabian tribe ancestry) at the juncture of the clusters of African, Asian, and European genomes; this is in congruence with the geographical location of the Arabian Peninsula. The Peninsula is at the nexus of Africa, Europe and Asia and has been implicated as part of early human migration route out of Africa [46], [47] and of early intercontinental trade routes [48]. The tree further illustrates that the global distribution of known disease-causing and predisposing variants within every genome is influenced by the individual's ethnicity. Through this study, we report analysis of a personal genome sequence from the contexts of both population genetics and genotype-phenotype associations. The reported reference data set of genome variants from the individual of “tent-dwelling” Bedouin ancestry from the Peninsula helps to enrich understanding of human genome variation across diverse populations. This is particularly important given that the large-scale global sequencing projects have not so far considered populations from Arabian Peninsula.
The rate of consanguineous mating in Kuwaiti population can be as high as 54.3% with higher rates noted among Bedouin tribes [45], [49], [50]. Frequency of intermarriage with other communities has been particularly low and this has resulted in sustained isolation particularly for the Bedouins and wealthy families. As a result of the extreme inbreeding, consanguinity, and isolation over many centuries, Bedouins (& other Arab tribes) exhibit a high incidence of genetic disorders [45], particularly autosomal recessive disorders. Studies with Bedouins in Negev region of Israel also have shown that they exhibit a high rate of genetically-determined neurological, skeletal, eye, cardiac, gastro-intestinal, skin and eye diseases [9]. Thus the Bedouin population is of considerable interest to the medical genetics community that strives to understand the pathophysiology of genetic disorders. Therefore, the reported full-length reference genome sequence for the “tent-dwelling” Bedouins is significantly important to the medical genetics community. Moreover, we find a large number of potentially deleterious missense variants (both known and novel), annotated for diseases in OMIM or GWAS Central catalog, which could be causal variants for a number of autosomal recessive disorders.
Examination of potentially deleterious missense SNPs from the reported genome for disease annotation (in OMIM and GWAS Catalog) leads to deciphering the relationship between the genotype and phenotype characteristics of the individual. (a) The participant is known to suffer from bronchial asthma. A potentially deleterious variant is seen at rs1051931 T = > C (A379V) in PLA2G7 gene; the A379V change has been shown to contribute to increased risk for asthma and atopy [33]. Asthma and related allergic diseases are complex conditions caused by a combination of genetic and environmental factors—the environmental factors differ from population to population. Kuwait, like other states of the Arabian Peninsula, has an arid climate with very hot dry summers and mild winters. Sandstorms are a regular climatic feature occurring most frequently in summer. Rapid urbanization in the post-oil era further contributes to increasing prevalence of asthma in Kuwait and other states. (b) The participant is morbid obese and has abnormal waist circumference. Potentially deleterious variants seen at the following markers have been associated with obesity and related traits (such as abdominal obesity): rs3733418 (C4orf39), rs2043112 (RICTOR), rs2275848 (NINJ1), rs11042023 (RPL27A), and rs1545 (MKKS) all carrying risk allele in the genome of the participant. Both adult obesity and childhood obesity are prevalent in Kuwait. (c) The participant has a family history of retinopathy. The individual carries risk allele at rs10151259 G = > T (A547S) (RPGRIP1) that is associated with Cone-rod dystrophy 13. (d) The participant is a smoker. Potentially deleterious variant rs1801272 A = > T (Leu160His) (CYP2A6) that is associated with smoking behavior is seen in the genome of the participant. Further, the study illustrates that the genome contains risk alleles for many autosomal disorders which are prevalent in the region. Examples include (a) Familial Mediterranean Fever (that can occur in both autosomal dominant and autosomal recessive forms)—marked by rs3743930 G➜ C (E148Q) (MEFV gene) in the sequenced Bedouin genome—affects predominantly populations living in the Mediterranean region, especially North African Jews, Armenians, Turks, and Arabs [51]; and (b) Parkinson disease (autosomal dominant) is marked by rs7133914 G = > A(R1398H) (LRRK2 gene) and rs7308720 C = > G (N551K) (again LRRK2 gene) in the sequenced Bedouin genome; while the LRRK2 mutations account for only about 1–2% of sporadic Parkinson's disease cases worldwide, in genetically isolated populations, such as Ashkenazi Jews and North African Arabs, the mutations can account for upwards of 30–40% of sporadic and familial PD cases [52].
Neighbor-joining trees, depicting comparisons between the genome of nomadic Bedouin (KWB) individual (along with two genomes from the KWS subgroup of Saudi Arabian tribe ancestry) and genomes from four continents, give different information depending on whether all the shared genome-wide SNPs or only those shared disease-associated SNPs (as cataloged in OMIM database) are used. While the neighbor-joining tree based on genome-wide SNPs clusters the KWB and KWS genomes amidst the European genomes, that based on only the OMIM SNPs places the KWB and KWS genomes between the three clusters of African, Asian and European genomes (in concordance with the geographical location of the origin of the sample at the nexus of Africa, Asia and Europe). This illustrates that the disease profile of individual populations can be different, irrespective of their overall shared origin, and that ethnicity acts as the dominant trend structuring disease-associated SNP locations. This is in agreement with reports that increased levels of population differentiation are detected in disease associated genes when compared to genome-wide base levels [53]. Placement of Kuwaiti genomes amidst the European genomes in the tree based on genome-wide SNPs is in concordance with the following reports: It has been recently suggested, based on analysis of ancient European genomes, that one of the three groups to which the present-day Europeans trace their ancestry is Middle Eastern farmers [54], [55]; the three groups are hunter–gatherers who arrived from Africa more than 40,000 years ago, Middle Eastern farmers who migrated to the west much more recently, and those that probably spanned between northern Europe and Siberia. The ancestry admixture due to Middle Eastern farmers in European ancestry may account, at least partially, for the affinity that we see between Europeans and the KWB & KWS participants in the tree based on genome-wide variants.
In conclusion, this is the first study to report a reference genome resource for the population of nomadic “tent-dwelling” Bedouin ancestry. We report novel genome variants that include SNPs, indels and structural variations that enlarge the current repertoire of human genome variation. Neighbor-joining tree built using shared disease-causing variants between the Bedouin genome and other continental genomes positions the Bedouin genome between the clusters of African and clusters of Asian and European genomes; this is in concordance with the geographical location of the origin of the sample at the nexus of Africa, Asia and Europe. Apart from the findings from population-context, the study illustrates that the medical history of the participant for morbid obesity and bronchial asthma as well as the medical history of the participant's family for retinopathy is accounted by the presence of a large number of genome variants that are known to be associated with these traits. Further, the study illustrates that the genome contains risk alleles for autosomal disorders that are prevalent in the region. The presented genome data provides a starting point for designing large-scale genetic studies in population subgroup of Bedouin ancestry in Kuwait and other states of the Middle East and North Africa.
Data availability
The reported whole genome sequence and all the identified variants (known and novel) are available on the ftp site (ftp://dgr.dasmaninstitute.org). The data can be visualized using genome browser with other annotations tracks from UCSC at http://dgr.dasmaninstitute.org/DGR/gb.html. Proper functionality of the web server requires Firefox version 6 (or later versions) or Internet Explorer version 10 (or later versions).
Materials and methods
Ethics statement
The study was approved by the Scientific Advisory Board and the Ethics Advisory Committee at Dasman Diabetes Institute, Kuwait. Written informed consent for the study was obtained from participant before blood samples were collected.
Detailed methodologies
Details on the methodologies and the tools used to process sample, to sequence the whole genome, and to analyze the genome and variants are presented in Supplementary Data—Appendix A. We present below only the essential information on methodologies.
Participant recruitment and sample collection
A 20 year old male participant, clustering with the genetically distinct Bedouin subgroup (KWB) of Kuwaiti population [2], was considered for whole genome sequencing. Blood sample was collected by a trained nurse. Ancestry estimates for the sequenced sample are as extracted from our previous study [2]. For purposes of illustrating the placement of this sample in the Bedouin genetic cluster, the principal component analysis (PCA) plot derived from our previous work [2] is used.
Whole genome sequencing
Processing of blood sample and preparation of libraries for whole genome sequencing were performed as per standard procedures. Paired-end sequencing was performed using Illumina HiSeq 2000.
Identification of genome variants (SNP and Indel) and validation of SNPs
Sequenced paired-end reads were aligned to human reference genome hg19 (UCSC) [56]. The aligned reads were processed to identify SNPs and indels using SAMtools [57] and GATK [58], [59] workflows; in order to reduce the likelihood of false discoveries due to the choice of the variant caller, we only utilized the consensus set of variants identified by both the tools. The validity of the SNP calls was confirmed by utilizing genome-wide genotype data from the same sample.
Annotation of variants (SNPs and indels)
A variant is denoted as “novel” if either the variant is not annotated in dbSNP 138 [17] database or the alternate allele seen in the variant in the sample is not a subset of alleles reported in dbSNP. SIFT [22] and PolyPhen2 [23] were used to annotate non-synonymous variants as “deleterious variants” depending on the predicted impact of the amino acid substitution on the protein functionality. The databases of OMIM [26], GWAS Catalog [25], and Ensembl Variation database v72 [60] were used to annotate variants for disease associations.
Detecting structural variations
We used HugeSeq [61] pipeline that implements four different algorithms, to detect structural variations from paired-end reads data. Deletions were annotated using Annovar [62]. A detected deletion is defined to be ‘known’ if at least 50% of the detected deletion overlaps with annotated deletions in the Database of Genomic Variants [43]; otherwise, the deletion is considered to be “novel”.
Identifying Y-chromosome and mitochondrial haplogroup
The Y-chromosome variants were used to call haplogroups using AMY-tree software [63], which uses data from ISOGG (International Society of Genetic Genealogy). The paired-end reads aligned to hg19 mitochondrial sequence were realigned to rCRS (Revised Cambridge Reference Sequence [64]) and then the variants were used to call haplogroups using HaploGrep software [65].
Neighbor-joining trees based on intergenome distances between the genomes of Bedouin, Saudi Arabian tribes, and continental populations
We consider a total of 10 genomes (downloaded from the sites of 10Gen [44]) covering diverse ethnicities from other continents, and 2 Kuwaiti genomes of Saudi Arabian tribe ancestry [10] for comparing the intergenome similarities with the genome of Bedouin ancestry sequenced in this study. The data set of 10 genomes from other continents includes three African (Yoruba) genomes (NA19240 [44], [66] on ABI SOLiD, NA18507 [67] on Illumina, NA18507 [68]) on ABI SOLiD), two Asian genomes (Chinese [69] on Illumina, Korean genome [70] on Illumina), five genomes of European descent (Venter [71] on Sanger sequencing, Watson [72] on Roche 454, NA07022 [73] on CGenomics, NA12878 [44], [66] on ABI SOLiD, Quake [74] on Helicos). The two Kuwaiti genomes of Saudi Arabian tribe ancestry and the third Kuwaiti genome of nomadic Bedouin ancestry are sequenced using Illumina sequencing technology. We adopt the methods used by Moore et al. [44] to calculate intergenome distances based on information relating to shared variant locations between genomes, and to create a consensus neighbor-joining tree (using PHYLIP [75]) depicting intergenome similarities. The method to calculate the distance is robust with respect to the depth of coverage and hence works well with genomes even when they are sequenced using different sequencing technologies. We built two trees: (i) genome-wide variant tree, that is based on intergenome distances calculated using genome-wide shared variant locations; (ii) OMIM variant tree, that is based on intergenome distances calculated using only those shared variant locations where at least one of the genomes contains an OMIM allele.
Visualization of the content of sequenced genome
The software tool, Circos [76] is used to create the high-level view of the contents (such as density of duplications) of the draft genome sequence. We have built a genome browser, using JBrowse (version 1.8.1) [77] to visualize the sequenced genome sequence and the variants.
Competing interests
The authors declare that they do not have any competing interests.
Author contributions
The study design was performed by OA, TAT and KB. OA directed sample collection & sequencing experiments and contributed to writing the manuscript. TAT directed the data analysis, and developed the manuscript. KB participated in discussions and approved the manuscript. GT and SEJ performed all the data analysis and contributed substantially to writing the manuscript. PH developed the data dissemination protocols and the web sites.
Acknowledgments
The authors thank Philip Beales and Mike Hubank (University College of London Genomics, London) for their advice and suggestions. The authors thank Antony Brooks (University College of London Genomics, London) for help with preparing sequencing libraries. The authors thank Dr Bahareh Azizi for her support and encouragement. The authors thank Daisy Thomas, Motasem K Melhem, Maisa Mahmoud and Ghazi Alghanim for help with recruiting participants. The Ethical Committee and the Scientific Advisory Board at Dasman Diabetes Institute are acknowledged for approving the study. The Kuwait Foundation for the Advancement of Sciences (KFAS) is acknowledged for funding the activities at our institute.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.gdata.2014.11.016.
Contributor Information
Thangavel Alphonse Thanaraj, Email: Alphonse.Thangavel@dasmaninstitute.org.
Osama Alsmadi, Email: osama.alsmadi@dasmaninstitute.org.
Appendix A. Supplementary data
Detailed Materials and Methods
Supplementary Tables
References
- 1.Casey M.S., Thackeray F.W., Findling J.E. Greenwood Press; Westport, Conn: 2007. The History of Kuwait. [Google Scholar]
- 2.Alsmadi O., Thareja G., Alkayal F., Rajagopalan R., John S.E., Hebbar P., Behbehani K., Thanaraj T.A. Genetic substructure of Kuwaiti population reveals migration history. PLoS One. 2013;8(9):e74913. doi: 10.1371/journal.pone.0074913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu-Amero K.K., Larruga J.M., Cabrera V.M., Gonzalez A.M. Mitochondrial DNA structure in the Arabian Peninsula. BMC Evol. Biol. 2008;8:45. doi: 10.1186/1471-2148-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu-Amero K.K., Hellani A., Gonzalez A.M., Larruga J.M., Cabrera V.M., Underhill P.A. Saudi Arabian Y-Chromosome diversity and its relationship with nearby regions. BMC Genet. 2009;10:59. doi: 10.1186/1471-2156-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richards M., Rengo C., Cruciani F., Gratrix F., Wilson J.F., Scozzari R., Macaulay V., Torroni A. Extensive female-mediated gene flow from sub-Saharan Africa into near eastern Arab populations. Am. J. Hum. Genet. 2003;72(4):1058–1064. doi: 10.1086/374384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter-Zinck H., Musharoff S., Salit J., Al-Ali K.A., Chouchane L., Gohar A., Matthews R., Butler M.W. Population genetic structure of the people of Qatar. Am. J. Hum. Genet. 2010;87(1):17–25. doi: 10.1016/j.ajhg.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hitti P.K. Regnery Publishing; Washington, D.C.: 1996. The Arabs: A Short History. [Google Scholar]
- 8.Chatty D. Brill. Netherlands; Boston: 2006. Nomadic societies in the Middle East and North Africa: entering the 21st century. [Google Scholar]
- 9.Markus B., Alshafee I., Birk O.S. Deciphering the fine-structure of tribal admixture in the Bedouin population using genomic data. Heredity (Edinburgh) 2014;112(2):182–189. doi: 10.1038/hdy.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alsmadi O., John S.E., Thareja G., Hebbar P., Antony D., Behbehani K., Thanaraj T.A. Genome at juncture of early human migration: a systematic analysis of two whole genomes and thirteen exomes from Kuwaiti population subgroup of inferred Saudi Arabian tribe ancestry. PLoS One. 2014;9(6):e99069. doi: 10.1371/journal.pone.0099069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thareja G., John S.E., Hebbar P., Behbehani K., Thanaraj T.A., Alsmadi O. Comprehensive analysis of a personal genome of Persian ancestry from Kuwait. BMC Genomics. 2015 doi: 10.1186/s12864-015-1233-x. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiaroni J., King R.J., Myres N.M., Henn B.M., Ducourneau A., Mitchell M.J., Boetsch G., Sheikha I. The emergence of Y-chromosome haplogroup J1e among Arabic-speaking populations. Eur. J. Hum. Genet. 2010;18(3):348–353. doi: 10.1038/ejhg.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behar D.M., Villems R., Soodyall H., Blue-Smith J., Pereira L., Metspalu E., Scozzari R., Makkan H. The dawn of human matrilineal diversity. Am. J. Hum. Genet. 2008;82(5):1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kivisild T., Reidla M., Metspalu E., Rosa A., Brehm A., Pennarun E., Parik J., Geberhiwot T. Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears. Am. J. Hum. Genet. 2004;75(5):752–770. doi: 10.1086/425161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fendt L., Rock A., Zimmermann B., Bodner M., Thye T., Tschentscher F., Owusu-Dabo E., Gobel T.M. MtDNA diversity of Ghana: a forensic and phylogeographic view. Forensic Sci. Int. Genet. 2012;6(2):244–249. doi: 10.1016/j.fsigen.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allard M.W., Polanskey D., Miller K., Wilson M.R., Monson K.L., Budowle B. Characterization of human control region sequences of the African American SWGDAM forensic mtDNA data set. Forensic Sci. Int. 2005;148(2–3):169–179. doi: 10.1016/j.forsciint.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenna K.P., McLaughlin R.L., Byrne S., Elamin M., Heverin M., Kenny E.M., Cormican P., Morris D.W. Delineating the genetic heterogeneity of ALS using targeted high-throughput sequencing. J. Med. Genet. 2013;50(11):776–783. doi: 10.1136/jmedgenet-2013-101795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez-Flores J.L., Fuller J., Hackett N.R., Salit J., Malek J.A., Al-Dous E., Chouchane L., Zirie M. Exome sequencing of only seven Qataris identifies potentially deleterious variants in the Qatari population. PLoS One. 2012;7(11):e47614. doi: 10.1371/journal.pone.0047614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong L.P., Ong R.T., Poh W.T., Liu X., Chen P., Li R., Lam K.K., Pillai N.E. Deep whole-genome sequencing of 100 southeast Asian Malays. Am. J. Hum. Genet. 2013;92(1):52–66. doi: 10.1016/j.ajhg.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 23.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKusick V.A. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007;80(4):588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Comuzzie A.G., Cole S.A., Laston S.L., Voruganti V.S., Haack K., Gibbs R.A., Butte N.F. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7(12):e51954. doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheeler E., Huang N., Bochukova E.G., Keogh J.M., Lindsay S., Garg S., Henning E., Blackburn H. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat. Genet. 2013;45(5):513–517. doi: 10.1038/ng.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berndt S.I., Gustafsson S., Magi R., Ganna A., Wheeler E., Feitosa M.F., Justice A.E., Monda K.L. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 2013;45(5):501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraja A.T., Vaidya D., Pankow J.S., Goodarzi M.O., Assimes T.L., Kullo I.J., Sovio U., Mathias R.A. A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes. 2011;60(4):1329–1339. doi: 10.2337/db10-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hotta K., Nakamura T., Takasaki J., Takahashi H., Takahashi A., Nakata Y., Kamohara S., Kotani K. Screening of 336 single-nucleotide polymorphisms in 85 obesity-related genes revealed McKusick–Kaufman syndrome gene variants are associated with metabolic syndrome. J. Hum. Genet. 2009;54(4):230–235. doi: 10.1038/jhg.2009.16. [DOI] [PubMed] [Google Scholar]
- 33.Kruse S., Mao X.Q., Heinzmann A., Blattmann S., Roberts M.H., Braun S., Gao P.S., Forster J. The Ile198Thr and Ala379Val variants of plasmatic PAF-acetylhydrolase impair catalytical activities and are associated with atopy and asthma. Am. J. Hum. Genet. 2000;66(5):1522–1530. doi: 10.1086/302901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirin M., Chandra A., Charteris D.G., Hayward C., Campbell S., Celap I., Bencic G., Vatavuk Z. Genome-wide association study identifies genetic risk underlying primary rhegmatogenous retinal detachment. Hum. Mol. Genet. 2013;22(15):3174–3185. doi: 10.1093/hmg/ddt169. [DOI] [PubMed] [Google Scholar]
- 35.Hameed A., Abid A., Aziz A., Ismail M., Mehdi S.Q., Khaliq S. Evidence of RPGRIP1 gene mutations associated with recessive cone-rod dystrophy. J. Med. Genet. 2003;40(8):616–619. doi: 10.1136/jmg.40.8.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorgeirsson T.E., Gudbjartsson D.F., Surakka I., Vink J.M., Amin N., Geller F., Sulem P., Rafnar T. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 2010;42(5):448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makela K.M., Seppala I., Hernesniemi J.A., Lyytikainen L.P., Oksala N., Kleber M.E., Scharnagl H., Grammer T.B. Genome-wide association study pinpoints a new functional apolipoprotein B variant influencing oxidized low-density lipoprotein levels but not cardiovascular events: AtheroRemo Consortium. Circ. Cardiovasc. Genet. 2013;6(1):73–81. doi: 10.1161/CIRCGENETICS.112.964965. [DOI] [PubMed] [Google Scholar]
- 38.Goodloe R., Brown-Gentry K., Gillani N.B., Jin H., Mayo P., Allen M., McClellan B., Jr., Boston J. Lipid trait-associated genetic variation is associated with gallstone disease in the diverse Third National Health and Nutrition Examination Survey (NHANES III) BMC Med. Genet. 2013;14:120. doi: 10.1186/1471-2350-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoh J., Ott J. Mathematical multi-locus approaches to localizing complex human trait genes. Nat. Rev. Genet. 2003;4(9):701–709. doi: 10.1038/nrg1155. [DOI] [PubMed] [Google Scholar]
- 40.McCarthy M.I., Hirschhorn J.N. Genome-wide association studies: potential next steps on a genetic journey. Hum. Mol. Genet. 2008;17(R2):R156–R165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chavarria-Soley G., Sticht H., Aklillu E., Ingelman-Sundberg M., Pasutto F., Reis A., Rautenstrauss B. Mutations in CYP1B1 cause primary congenital glaucoma by reduction of either activity or abundance of the enzyme. Hum. Mutat. 2008;29(9):1147–1153. doi: 10.1002/humu.20786. [DOI] [PubMed] [Google Scholar]
- 42.Vreken P., Van Kuilenburg A.B., Meinsma R., van Gennip A.H. Identification of novel point mutations in the dihydropyrimidine dehydrogenase gene. J. Inherit. Metab. Dis. 1997;20(3):335–338. doi: 10.1023/a:1005357307122. [DOI] [PubMed] [Google Scholar]
- 43.MacDonald J.R., Ziman R., Yuen R.K., Feuk L., Scherer S.W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42(Database issue):D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore B., Hu H., Singleton M., De La Vega F.M., Reese M.G., Yandell M. Global analysis of disease-related DNA sequence variation in 10 healthy individuals: implications for whole genome-based clinical diagnostics. Genet. Med. 2011;13(3):210–217. doi: 10.1097/GIM.0b013e31820ed321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teebi A.S. Autosomal recessive disorders among Arabs: an overview from Kuwait. J. Med. Genet. 1994;31(3):224–233. doi: 10.1136/jmg.31.3.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cabrera V., Abu-Amero K., Larruga J., González A. The Arabian Peninsula: Gate for Human Migrations Out of Africa or Cul-de-Sac? A Mitochondrial DNA Phylogeographic Perspective. In: Petraglia M.D., Rose J.I., editors. The Evolution of Human Populations in Arabia. Springer; Netherlands: 2010. pp. 79–87. [Google Scholar]
- 47.Rose J., Petraglia M. Tracking the Origin and Evolution of Human Populations in Arabia. In: Petraglia M.D., Rose J.I., editors. The Evolution of Human Populations in Arabia. Springer; Netherlands: 2010. pp. 1–12. [Google Scholar]
- 48.Slot B. Arabian Publishing; London: 2003. Kuwait: The Growth of a Historic Identity. [Google Scholar]
- 49.Al-Awadi S.A., Moussa M.A., Naguib K.K., Farag T.I., Teebi A.S., el-Khalifa M., el-Dossary L. Consanguinity among the Kuwaiti population. Clin. Genet. 1985;27(5):483–486. doi: 10.1111/j.1399-0004.1985.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 50.Al-Nassar K.E., Kelly C.L., EL-Kazimi A. Patterns of consanguinity in the population of Kuwait. Am. J. Hum. Genet. 1989;45(Suppl. 4):0915A. [Google Scholar]
- 51.Shohat M., Halpern G.J. Familial Mediterranean fever—a review. Genet. Med. 2011;13(6):487–498. doi: 10.1097/GIM.0b013e3182060456. [DOI] [PubMed] [Google Scholar]
- 52.Haas B.R., Stewart T.H., Zhang J. Premotor biomarkers for Parkinson's disease—a promising direction of research. Transl. Neurodegener. 2012;1(1):11–23. doi: 10.1186/2047-9158-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amato R., Pinelli M., Monticelli A., Marino D., Miele G., Cocozza S. Genome-wide scan for signatures of human population differentiation and their relationship with natural selection, functional pathways and diseases. PLoS One. 2009;4(11):e7927. doi: 10.1371/journal.pone.0007927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Callaway, E., Ancient European genomes reveal jumbled ancestry: Nature News & Comment. DOI: citeulike-article-id:12904863.
- 55.Lazaridis I., Patterson N., Mittnik A., Renaud G., Mallick S., Kirsanow K., Sudmant P.H., Schraiber J.G. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513(7518):409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 57.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van der Auwera G.A., Carneiro M., Hartl C., Poplin R., del Angel G., Levy-Moonshine A., Jordan T., Shakir K. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Current Protocols in Bioinformatics. 2013;43 doi: 10.1002/0471250953.bi1110s43. 11.10.1-11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Flicek P., Ahmed I., Amode M.R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P. Ensembl 2013. Nucleic Acids Res. 2013;41(Database issue):D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lam H.Y., Pan C., Clark M.J., Lacroute P., Chen R., Haraksingh R., O'Huallachain M., Gerstein M.B. Detecting and annotating genetic variations using the HugeSeq pipeline. Nat. Biotechnol. 2012;30(3):226–229. doi: 10.1038/nbt.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Geystelen A., Decorte R., Larmuseau M.H. AMY-tree: an algorithm to use whole genome SNP calling for Y chromosomal phylogenetic applications. BMC Genomics. 2013;14:101. doi: 10.1186/1471-2164-14-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23(2):147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 65.Kloss-Brandstatter A., Pacher D., Schonherr S., Weissensteiner H., Binna R., Specht G., Kronenberg F. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum. Mutat. 2011;32(1):25–32. doi: 10.1002/humu.21382. [DOI] [PubMed] [Google Scholar]
- 66.De La Vega F.M., Hyland F.C.L., McLaughlin S., MacBride A.R., Tsung E.F., Peckham H., Scafe C., Lee C. 2009. Functional analysis of the genetic variation within the genomes of three HapMap individuals obtained by whole-genome, second-generation sequencing. ( https://tools.lifetechnologies.com/content/sfs/posters/cms_065553.pdf) [Google Scholar]
- 67.Bentley D.R., Balasubramanian S., Swerdlow H.P., Smith G.P., Milton J., Brown C.G., Hall K.P., Evers D.J. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McKernan K.J., Peckham H.E., Costa G.L., McLaughlin S.F., Fu Y., Tsung E.F., Clouser C.R., Duncan C. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19(9):1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J., Wang W., Li R., Li Y., Tian G., Goodman L., Fan W., Zhang J. The diploid genome sequence of an Asian individual. Nature. 2008;456(7218):60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ahn S.M., Kim T.H., Lee S., Kim D., Ghang H., Kim D.S., Kim B.C., Kim S.Y. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19(9):1622–1629. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Levy S., Sutton G., Ng P.C., Feuk L., Halpern A.L., Walenz B.P., Axelrod N., Huang J. The diploid genome sequence of an individual human. PLoS Biol. 2007;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wheeler D.A., Srinivasan M., Egholm M., Shen Y., Chen L., McGuire A., He W., Chen Y.J. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452(7189):872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 73.Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327(5961):78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 74.Pushkarev D., Neff N.F., Quake S.R. Single-molecule sequencing of an individual human genome. Nat. Biotechnol. 2009;27(9):847–850. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. (DOI: citeulike-article-id:2344765. [Google Scholar]
- 76.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Westesson O., Skinner M., Holmes I. Visualizing next-generation sequencing data with JBrowse. Brief. Bioinform. 2013;14(2):172–177. doi: 10.1093/bib/bbr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed Materials and Methods
Supplementary Tables
Data Availability Statement
The reported whole genome sequence and all the identified variants (known and novel) are available on the ftp site (ftp://dgr.dasmaninstitute.org). The data can be visualized using genome browser with other annotations tracks from UCSC at http://dgr.dasmaninstitute.org/DGR/gb.html. Proper functionality of the web server requires Firefox version 6 (or later versions) or Internet Explorer version 10 (or later versions).