Abstract
Colorectal cancer (CRC), which has high prevalence in Saudi Arabia and worldwide, needs better understanding by exploiting the latest available cytogenetic microarrays. We used biopsy tissue from consenting colorectal cancer patients to extract DNA and carry out microarray analysis using a CytoScan HD platform from Affymetrix. Patient specific comparisons of tumor–normal pairs were carried out. To find out the high probability key players, we performed Genomic Identification of Significant Targets in Cancer analysis and found 144 genes to form the list of driver genes. Of these, 24 genes attained high GISTIC scores and suggest being significantly associated with CRC. Loss of heterozygosity and uniparental disomy were found to affect 9 genes and suggest different mechanisms associated with CRC in every patient. Here we present the details of the methods used in carrying out the above analyses. Also, we provide some additional data on biomarker analysis that would complement the findings.
Keywords: Microarray, Colorectal cancer, CytoScan HD, GISTIC, Nexus
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens |
| Strain | Patient's colorectal tumor and adjacent normalmucosa |
| Sex | Both male and female |
| Array type | Affymetrix CytoScan HD |
| Data format | Raw data: CEL files, processed data: Excel table |
| Experimental factors | Tumor vs. normal |
| Experimental features | Tumor and normal samples compared for copy number aberrations, loss of heterozygosity, uniparental disomy, GISTIC analysis miRNA target prediction, effect on transcription factor binding sites, functional analysis, pathway and network analysis, biomarker analysis |
| Sample source location | National Guard Hospital, Riyadh, Saudi Arabia |
| Consent | All patients consented before starting the study |
Direct link to deposited data
Deposited data can be found at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47204.
Experimental design, materials and methods
We carried out patient specific comparison of tumor and normal samples. Each patient's tumor microarray profile was compared against its own normal profile. Copy number variation, loss of heterozygosity and uniparental disomy were studied and the affected genes were analyzed.
Sample collection
Biopsies were collected from Saudi Arabian patients presenting for preliminary CRC diagnosis. All cases were collected regardless of surgical stage or histologic grade. Each hematoxylin and eosin (H & E) stained case was reviewed by a board-certified pathologist to confirm the specimen's histological consistency with colon adenocarcinoma and that adjacent normal specimen contained no tumor cells. The sections were required to contain > 60% tumor cell nuclei for inclusion in the study. The cohort consisted of patients who have not undergone any known CRC-related clinical intervention prior to the time of biopsy acquisition. Multiple biopsies were taken from the normal and tumor tissues. All the patient samples were collected from a single hospital.
Sample processing & DNA extraction
Paired samples of tumor and adjacent normal mucosa taken from > 2 cm apart were collected. Each tumor specimen weighed between 10 and 30 mg. The biopsy tissue was stored in RNAlater (Ambion) at 4 °C for 24 h, followed by freezing and further storage at − 20 °C [1]. CRC-positive sample pairs were then selected for DNA extraction by NucleoSpin Trio Kit (Macherey-Nagel, Germany) [2]. 10–30 mg tissue was homogenized using TissueLyser and stainless steel beads from Qiagen. Quality and quantity checks were carried out by Nanodrop (Thermo Fischer Scientific).
Data generation using cytogenetic array
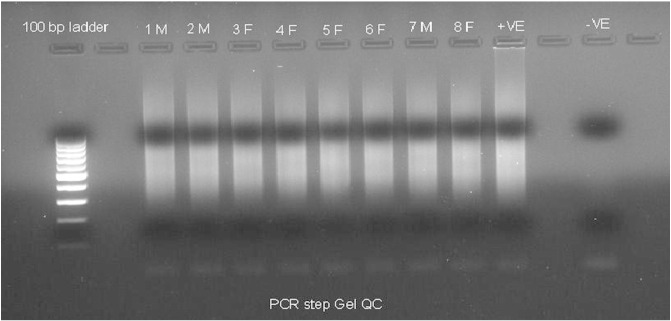
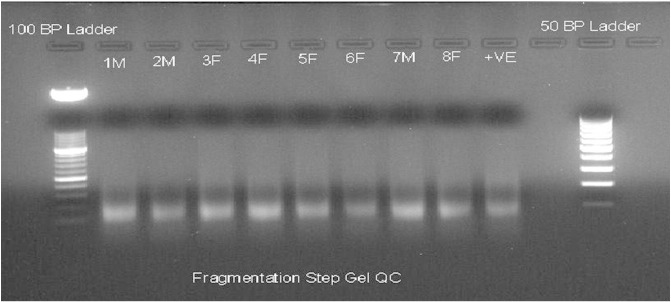
CytoScan HD arrays along with a complete kit were acquired from Affymetrix (Affymetrix Inc., USA). The recommended DNA amplification kit was obtained from Clontech (Clontech Laboratories Inc., USA). The supplier's protocol was followed for the amplification, hybridization, washing and staining steps. Quality control after PCR amplification (Fig. 1) and fragmentation (Fig. 2) was carried out using agarose gel electrophoresis. The arrays were scanned using a 30007G scanner from Affymetrix.
Fig. 1.
Agarose gel electrophoresis of PCR amplified DNA. Quality control measure for checking amplified DNA requires running agarose gel. Representative samples from patient samples with IDs 1M–8F (as mentioned in the original manuscript [3]) are provided here along with the positive sample as supplied in the CytoScan HD kit. Negative control is just water in place of DNA.
Fig. 2.
Agarose gel electrophoresis of DNA after fragmentation step. Quality control for DNA after fragmentation step. Representative samples from patient samples with IDs 1M–8F (as mentioned in the original manuscript [3]) are provided here along with the positive sample as supplied in the CytoScan HD kit. Negative control is just water in place of DNA.
Data analysis
We followed a case–control analysis strategy where the subject served as the donor of both control and tumor tissues. The tumor–normal comparisons were thus carried out between homogenous samples.
CNV, LOH and UPD analyses
Nexus Copy Number 6.0 (Biodiscovery, Inc., CA, USA) was used to assess genome wide copy number frequencies for the 15 patients. Furthermore, Aroma.affymetrix – another CBS implementation as part of the BioConductor's DNACopy library and the associated TumorBoost algorithm (which normalize allele specific copy numbers for tumor samples with paired normal) – was also used to identify genomic events. We obtained conforming results from both implementations. The combination of CytoScan HD's high resolution and in depth analysis by Nexus Copy Number allows us to capture even the smallest genomic events. All the samples had a quality score of < 0.2 (Table 1). The frequency threshold parameters used for the analysis are 0.2 for gain, 0.6 for high gain, − 0.2 for slight loss and − 1.0 for big loss. A minimum cutoff of 500 kb was used for detecting these events.
Table 1.
Summary of processed samples.
| Sample | Data type | Quality | One copy gain | One copy loss | Two or more copy gain | Two copy loss | LOH | Total CN aberrations |
|---|---|---|---|---|---|---|---|---|
| 1M | Affymetrix CEL | 0.060 | 8 | 12 | 0 | 0 | 0 | 20 |
| 2M | Affymetrix CEL | 0.105 | 38 | 42 | 2 | 0 | 2 | 82 |
| 3F | Affymetrix CEL | 0.151 | 15 | 10 | 0 | 0 | 0 | 25 |
| 4F | Affymetrix CEL | 0.096 | 8 | 10 | 0 | 0 | 0 | 18 |
| 5F | Affymetrix CEL | 0.169 | 7 | 7 | 2 | 0 | 0 | 16 |
| 6F | Affymetrix CEL | 0.148 | 19 | 22 | 10 | 7 | 9 | 58 |
| 7M | Affymetrix CEL | 0.126 | 28 | 48 | 0 | 1 | 2 | 77 |
| 8F | Affymetrix CEL | 0.071 | 6 | 3 | 0 | 0 | 1 | 9 |
| 9F | Affymetrix CEL | 0.132 | 27 | 27 | 1 | 1 | 4 | 56 |
| 10F | Affymetrix CEL | 0.087 | 34 | 49 | 14 | 7 | 7 | 104 |
| 11F | Affymetrix CEL | 0.105 | 9 | 11 | 0 | 0 | 0 | 20 |
| 12M | Affymetrix CEL | 0.113 | 28 | 19 | 8 | 1 | 1 | 56 |
| 13M | Affymetrix CEL | 0.085 | 5 | 12 | 0 | 0 | 0 | 17 |
| 14F | Affymetrix CEL | 0.174 | 49 | 16 | 3 | 0 | 0 | 68 |
| 15M | Affymetrix CEL | 0.105 | 9 | 4 | 3 | 0 | 1 | 16 |
Information regarding transcription factor binding sites was obtained from the Open Regulatory Annotation Database (ORegAnno, www.oreganno.org).
miRNA target analysis was carried out using the microRNA integration system for target gene prediction (MIRSYSTEM) software version 20130328 available at http://mirsystem.cgm.ntu.edu.tw/.
GISTIC analysis
Combining GISTIC (Genomic Identification of Significant Targets in Cancer) score ranking and peaks in copy numbers of genomic regions, we identified the genes according to the annotation of the human genome assembly GRCh37/hg19.
Through Nexus Copy Number, we carried out GISTIC analysis. G-scores relay the significance of genes to drive cancer by weighing regions of aberration against the likelihood for random occurrence. The G-scores for regions detected by CBS were examined. We labeled as significant any region of a score above 2.
Biomarker discovery
We analyzed 144 genes identified through GISTIC for their potential role as biomarkers in human cancer. 31 genes were identified as potential biomarkers (Table 2). The full table of biomarker analysis with information about expression in body fluids is available as Supporting Table 1.
Table 2.
Biomarker molecules among the driver genes.
| Biomarker gene | Biomarker application |
|---|---|
| ADAM12 | Diagnosis |
| AGR3 | Diagnosis |
| AURKA | Efficacy |
| BCL2 | Diagnosis, efficacy, prognosis |
| CLDN7 | Unspecified application |
| CRP | Diagnosis, disease progression, efficacy, prognosis, safety |
| CYP19A1 | Diagnosis, efficacy |
| DCC | Prognosis |
| EXO1 | Diagnosis |
| FGFR2 | Diagnosis, response to therapy |
| GSTM1 | Diagnosis, efficacy, safety |
| HLTF | Diagnosis |
| ICAM1 | Diagnosis, efficacy, prognosis |
| IDO1 | Disease progression, efficacy, prognosis |
| IGF2BP3 | Diagnosis |
| IL6 | Diagnosis, disease progression, efficacy, prognosis, response to therapy, safety |
| IL6R | Efficacy |
| INHBA | Diagnosis |
| INSR | Prognosis |
| MGMT | Diagnosis, efficacy, prognosis |
| MS4A1 | Efficacy, prognosis |
| MUC4 | Diagnosis, disease progression |
| OVGP1 | Diagnosis |
| PTGS1 | Efficacy |
| PTK2 | Diagnosis, disease progression, efficacy, prognosis |
| PTP4A3 | Disease progression |
| SLC16A1 | Diagnosis |
| SMAD4 | Prognosis |
| STK11 | Diagnosis |
| TP53 | Diagnosis, disease progression, efficacy, prognosis, response to therapy |
| ZNF217 | Prognosis |
References
- 1.Life technologies “Storage of tissue over extended periods prior to RNA extraction can degrade RNA resulting in altered gene expression patterns”. http://www.lifetechnologies.com/sa/en/home/references/ambion-tech-support/rna-isolation/tech-notes/rna-remains-stable-during-long-term-tissue-storage.html Technotes available at. (as of 20 Feb., 2014)
- 2.Macherey-Nagel “Nucleospin trip kit”. http://www.mn-net.com/tabid/11113/default.aspx (as of 20 Feb., 2014)
- 3.Eldai H., Periysamy S., Alqarni S., Alrodayyan M., Mustafa S., Deeb A., Alsheikh E., Khan M.A., Yousef Z., Johani M., Aziz M.A. PLoS ONE. 2013;8(10):e76251. doi: 10.1371/journal.pone.0076251. [DOI] [PMC free article] [PubMed] [Google Scholar]