Abstract
Cardiovascular disease is a major cause of death worldwide, so people are advised to limit their intake of dietary cholesterol [1]. Egg consumption has been seriously reduced because of the high levels of cholesterol [2]. The objective of this study was to evaluate the cholesterol metabolism effects of alfalfa saponin extract (ASE) in liver and ovary tissues using digital gene-expression (DGE) profiling analysis. The liver and ovary tissues were isolated from laying hens fed with ASE for RNA sequencing. Here, we provide detailed experimental methods and analysis pipeline in our study to identify digital gene expression of alfalfa saponin extract on laying hens and analysis pipeline published by Singh and colleagues in the PLOS ONE [3]. The data generated in our work provide meaningful information for understanding the molecular mechanisms underlying the cholesterol-lowering effects of ASE.
Keywords: Digital gene-expression, Alfalfa saponin, Laying hens
| Specifications | |
|---|---|
| Organism/cell line/tissue | The liver and ovary tissues of laying hens (Hy-Line Brown hens) |
| Sex | Female |
| Sequencer or array type | Illumina Hiseq 2000 |
| Data format | Raw data: FASTQ files |
| Experimental factors | Fed with ASE based on basal diet vs. a basal diet with no ASE |
| Experimental features | The liver and ovary tissues of laying hens were collected and digital gene expression (DGE) was performed to investigate the mechanisms underlying the cholesterol-lowering effect of ASE. |
| Consent | N/A |
| Sample source location | All animal experiments were approved by the Animal Care and Use Committee of Henan Agricultural University. ASE was provided by Hebei Bao'en Biotechnology Co., Ltd. (Shijiazhuang, China) |
Direct link to deposited data
Deposited data can be found here:
Experimental design, materials and methods
Experimental design
Cardiovascular disease is a major cause of death worldwide, so people are advised to limit their intake of dietary cholesterol [1]. Egg consumption has been seriously reduced because of the high levels of cholesterol [2]. ASE can modulate cholesterol metabolism and decrease cholesterol levels in animals. In this study, the liver and ovary tissues of laying hens were collected and digital gene expression (DGE) was performed to investigate the mechanisms underlying the cholesterol-lowering effect of ASE. 150 Hy-Line Brown hens at the age of 27 weeks were randomly divided into five groups (five replicates were established per treatment, with 6 hens each replicate). Hens in each treatment were randomly fed five different experimental diets: Controls were fed a basal diet with 0 mg/kg ASE, and the other four groups were fed otherwise identical diets supplemented with 60 mg/kg, 120 mg/kg, 240 mg/kg, and 480 mg/kg ASE, respectively. The basal diet was designed to meet, or exceed, the National Research Council (NRC) nutrient requirements. The ingredients and calculated nutrient level of the basal diet are shown in Table 1 [3]. ASE was provided by Hebei Bao'en Biotechnology Co., Ltd. (Shijiazhuang, China) as a standardized product that contained 61.64% saponins, 10.97% flavonoids, 8.12% polysaccharides, 7.11% moisture, and 12.16% unknown compounds. ASE is extracted from Medicago sativa L. The main active component is saponins. And ASE played a positive role in lowering yolk cholesterol content [4], [5], [6]. According to the cholesterol levels in yolk of feeding experiment, the control group and 120 mg/kg ASE group were chosen for digital gene-expression profiling analysis (Table 2) [3]. The liver and ovary tissues of 120 mg/kg ASE group were isolated, frozen immediately in liquid nitrogen and stored at − 80 °C until RNA extraction (the liver from the control group (Li_CK), the liver from the 120 mg/kg ASE group (Li_Exp), the ovaries from the control group (Ov_CK), the ovaries from the 120 mg/kg ASE group (Ov_Exp)).
Table 1.
Composition and nutrient content of diet.
| Ingredient | % | Nutrient levelb | |
|---|---|---|---|
| Corn | 69.70 | Crude protein (%) | 16.50 |
| Soybean meal | 14.45 | Calcium (%) | 3.44 |
| Limestone power | 8.50 | Phosphorus (%) | 0.59 |
| Peruvian fishmeal | 4.96 | Available phosphorus (%) | 0.42 |
| Dicalcium phosphate | 1.00 | Lysine (%) | 0.83 |
| Salt | 0.20 | Methionine (%) | 0.38 |
| Choline chloride | 0.10 | Methionine + cysteine (%) | 0.63 |
| DL-methionine | 0.09 | Apparent metabolic energy (MJ/kg) | 11.49 |
| Trace mineral and vitamin premixa | 1.00 |
Premix provided the following per kilogram of diet: 60 mg of iron, 80 mg of manganese, 8 mg of copper, 80 mg of zinc, 1 mg of iodine, 0.3 mg of selenium, 12,200 IU of vitamin A, 4125 IU of vitamin D3, 30 IU of vitamin E, 4.5 mg of vitamin K, 5 mg of vitamin B12, 2 mg of biotin, 5 mg of folic acid, 32.5 mg of niacin, 5.3 mg of pantothenic acid, 8 mg of pyridoxine, 8.5 mg of riboflavin, and 1 mg of thiamine.
Obtained by calculation.
Table 2.
Dietary ASE and concentrations of yolk cholesterol (mg/g).
| Parameter period | ASE in diet (mg/kg) |
||||
|---|---|---|---|---|---|
| 0 | 60 | 120 | 240 | 480 | |
| Day 15 | 8.89 ± 0.21 | 8.20 ± 0.41 | 8.27 ± 0.07 | 8.46 ± 0.31 | 8.49 ± 0.19 |
| Day 30 | 9.23 ± 0.24a | 8.22 ± 0.25b | 8.35 ± 0.59b | 8.49 ± 0.61b | 8.49 ± 0.13b |
| Day 45 | 9.33 ± 0.24a | 9.09 ± 0.26ab | 8.78 ± 0.32b | 8.87 ± 0.11b | 9.02 ± 0.46ab |
| Day 60 | 9.57 ± 0.41a | 9.37 ± 0.25ab | 8.89 ± 0.45b | 9.47 ± 0.39ab | 9.31 ± 0.36ab |
a,bMeans in the same row not sharing a common superscript differ significantly at P < 0.05.
Total RNA isolation
Total RNA was isolated using the Trizol reagent (Invitrogen, Code No.: 15596-018) according to the manufacturer's instructions. RNA degradation and contamination were monitored on 1% agarose gels. RNA was assessed for quantity and quality using a Nanodrop ND-1000 spectrophotometer and Agilent 2100 Bioanalyzer according to the manufacturer's instructions. Contaminating genomic DNA in total RNA samples were treated with DNase-I.
Generation of sequencing data
2 μg of total RNA (≥ 100 ng/μL) was pooled from three tissues within each group: Li_CK, Li_Exp, Ov_CK, Ov_Exp. Four sequencing libraries were generated using NEB Next Ultra Directional RNA Library Prep Kit for Illumina (NEB) in accordance with the manufacturer's recommendations. MRNA was purified from total RNA using poly-T oligo-attached magnetic beads (Life technologies, CA, USA). Fragmentation was carried out using divalent captions under elevated temperature in NEB proprietary fragmentation buffer. First strand cDNA was synthesized using random oligonucleotides and M-Mel Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities and enzymes were removed. After adenylation of 3′ ends of DNA fragments, NEB Next adaptor oligonucleotides were ligated to cDNA fragments. In order to select cDNA fragments. DNA fragments with ligated adaptor molecules on both ends were selectively enriched using NEB Universal PCR Primer and Index primer in a 10 cycles PCR reaction. Products were purified and quantified using the Agilent high sensitivity DNA assay on the Agilent Bioanalyzer 2100 system. After the clustering of the index-coded samples, each library preparation was sequenced on an Illumina HiSeq 2000 platform and 50 bp single-end (SE) reads were generated.
Mapping and analysis of DGE reads
For each DGE library, clean tags were obtained by filtering out adaptor sequences and low-quality tags. Then the clean tags were retained and mapped to the reference genome of Gallus gallus at ftp://ftp.ensembl.org/pub/release-72/fasta/gallus_gallus/dna by TopHat v1.4.0 [7]. HTSeq v0.5.3 was used to count the reads mapped to each gene [8]. Then reads per kilobase transcriptome per million mapped reads (RPKM) of each gene were calculated based on the length of the gene and reads count mapped to this gene. It is currently the most common method of for estimating levels of gene expression [9]. In order to balance the number of false positives and false negatives a RPKM threshold value of 0.3 was established to determine whether or not given genes was expressed, as in several other studies [10], [11], [12]. Differential expression analysis of two conditions was performed using the DEGSeq R package v1.12.0 [13]. P-values were adjusted using Q-value. [14]. The Q-values of 0.05 and log2 (Fold_change) with no limitations were served as the threshold of significance for differential expression. Here, 110 significantly expressed genes were identified in liver samples; 66 were up-regulated and 44 were down-regulated. In ovary samples, 107 significantly expressed genes were obtained; 63 were up-regulated and 44 were down-regulated (Table S1) [3]. Gene Ontology (GO) enrichment analysis of differentially expressed genes was implemented by the GOseq R package [15]. We used KOBAS software to test the statistical enrichment of differential expression genes in KEGG pathways [16], [17]. KEGG pathway analysis of differently expressed genes was also performed (Table S2) [3]. Protein interaction networks of differentially expressed genes were acquired based on the information in STRING database (http://string-db.org/). Cytoscape software was used to visualize the interaction networks [18]. The process of DGE analysis was depicted in Fig. 1.
Fig. 1.
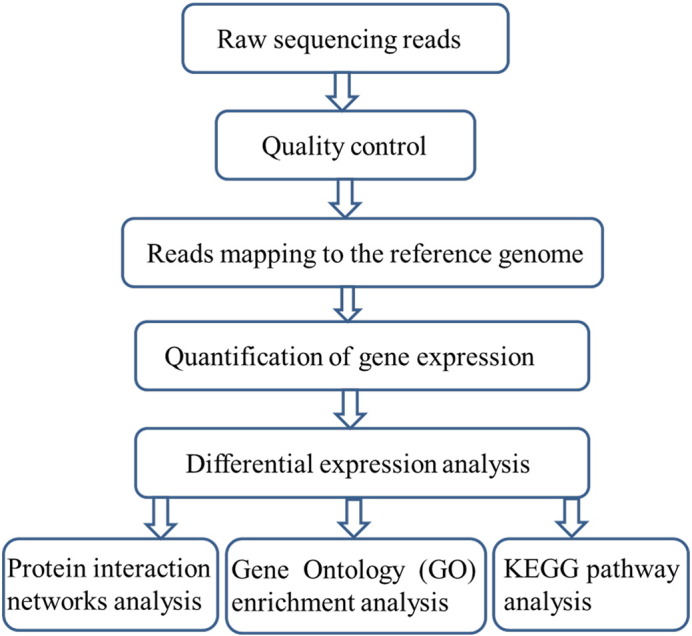
The process of DGE analysis.
Discussion
Based on the supplementing 120 mg/kg ASE played a positive role in lowering yolk cholesterol content [3], the group with 120 mg/kg ASE was used for digital gene-expression profiling analysis. In this study, some genes were found to be associated with the cholesterol metabolism. In order to shed more light on the functional roles of differentially expressed genes responsible for the cholesterol-lowering effect of ASE, functional annotation was investigated using GO classification and KEGG pathway analysis between Ov_CK and Ov_Exp samples [3]. Our results provided that ASE were partially mediated by enhancement of cholesterol efflux in the liver and this reduced of cholesterol deposition in the ovary.
The following are the Supplementary data related to this article.
Genes differentially expressed in the liver and ovary in ASE and control groups.
KEGG pathway classification of differentially expressed genes in the liver and the ovary.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
This work was funded by the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-35).
Contributor Information
Wenna Fan, Email: chou0516@163.com.
Hongqi Du, Email: biodhq@126.com.
Lu Zhou, Email: zhoulu06@gmail.com.
Pengfei Shi, Email: bird5682850@163.com.
Chengzhang Wang, Email: wangcz@henau.edu.cn.
References
- 1.Lung N.H., Institute B. Third report of the National Cholesterol Education Program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III): final report. Circulation. 2002;106:3143. [PubMed] [Google Scholar]
- 2.Spence J.D., Jenkins D.J., Davignon J. Egg yolk consumption and carotid plaque. Atherosclerosis. 2012;224:469–473. doi: 10.1016/j.atherosclerosis.2012.07.032. [DOI] [PubMed] [Google Scholar]
- 3.Zhou L., Shi Y., Guo R., Liang M., Zhu X., Wang C. Digital gene-expression profiling analysis of the cholesterol-lowering effects of alfalfa saponin extract on laying hens. PLoS ONE. 2014;9:e98578. doi: 10.1371/journal.pone.0098578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malinow M., McLaughlin P., Papworth L., Stafford C., Kohler G., Livingston A., Cheeke P. Effect of alfalfa saponins on intestinal cholesterol absorption in rats. Am. J. Clin. Nutr. 1977;30:2061–2067. doi: 10.1093/ajcn/30.12.2061. [DOI] [PubMed] [Google Scholar]
- 5.Malinow M., Connor W., McLaughlin P., Stafford C., Lin D., Livingston A., Kohler G., McNulty W. Cholesterol and bile acid balance in Macaca fascicularis. Effects of alfalfa saponins. J. Clin. Invest. 1981;67:156. doi: 10.1172/JCI110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu C.-H., Xie G., He R.-R., Zhai Y.-J., Li Y.-F., Tsoi B., Kurihara H., Yang D.-P. Effects of a purified saponin mixture from alfalfa on plasma lipid metabolism in hyperlipidemic mice. J. Health Sci. 2011;57:401–405. [Google Scholar]
- 7.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anders S. HTSeq: analysing high-throughput sequencing data with Python. 2010. http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html [DOI] [PMC free article] [PubMed]
- 9.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 10.Beyer M., Mallmann M.R., Xue J., Staratschek-Jox A., Vorholt D., Krebs W., Sommer D., Sander J., Mertens C., Nino-Castro A. High-resolution transcriptome of human macrophages. PLoS ONE. 2012;7:e45466. doi: 10.1371/journal.pone.0045466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sam L.T., Lipson D., Raz T., Cao X., Thompson J., Milos P.M., Robinson D., Chinnaiyan A.M., Kumar-Sinha C., Maher C.A. A comparison of single molecule and amplification based sequencing of cancer transcriptomes. PLoS ONE. 2011;6:e17305. doi: 10.1371/journal.pone.0017305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright H.L., Thomas H.B., Moots R.J., Edwards S.W. RNA-Seq reveals activation of both common and cytokine-specific pathways following neutrophil priming. PLoS ONE. 2013;8:e58598. doi: 10.1371/journal.pone.0058598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storey J.D. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 2003:2013–2035. [Google Scholar]
- 15.Young M., Wakefield M., Smyth G., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao X., Cai T., Olyarchuk J.G., Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes differentially expressed in the liver and ovary in ASE and control groups.
KEGG pathway classification of differentially expressed genes in the liver and the ovary.


