Abstract
BACKGROUND
Fluorescence in situ hybridization can detect genomic abnormalities in up to 80% of cases and provides prognostic information on patients with chronic lymphocytic leukemia (CLL). Although 13q deletion as the sole abnormality has been found to confer a favorable prognosis, there are little data as to whether there is a difference in prognostic value between monoallelic versus biallelic deletion of 13q.
METHODS
The authors reviewed the electronic database for patients with CLL who carried the 13q deletion as the sole abnormality and presented to The University of Texas MD Anderson Cancer Center (MDACC). Untreated patients were separated into 2 groups: those having monoallelic versus those with biallelic deletion of 13q. Using Mann-Whitney, chi-square, and Kaplan-Meier analysis, the baseline quantitative and qualitative variables for each group, along with the time from presentation to MDACC to treatment, were compared.
RESULTS
A total of 176 patients were identified; 143 patients had a monoallelic deletion of 13q, whereas 33 patients had a biallelic deletion. The only significantly different values between the groups were albumin (4.5 g/dL vs 4.4 g/dL; P = .01) and zeta-chain-associated protein kinase 70 (ZAP70) expression (1.7% vs 4.8%; P = .010). The median time from fluorescence in situ hybridization analysis to treatment in both the monoallelic and biallelic groups had not been reached (P = not significant).
CONCLUSIONS
Except for inconsequential differences in albumin and ZAP70 expression, there was no difference in the baseline characteristics between patients with CLL who had monoallelic or biallelic deletion of 13q. In addition, there was no significant difference in endpoints, including time to treatment.
Keywords: monoallelic, biallelic, 13q, chronic lymphocytic leukemia, prognostics
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the Western world and affects predominantly older adults. The clinical course and biology of this disease are heterogeneous; some patients die within months of diagnosis, whereas others live longer than 20 years.1 The Rai and Binet staging systems help identify patients who have more advanced disease and require treatment.2 Because most patients present with early- or intermediate-stage disease, additional markers are needed to stratify patients who are at increased risk of disease progression with concomitant decreased survival. Chromosome abnormalities provide prognostic information in CLL.3 The prognostic impact of cytogenetics on survival is underscored by a median survival of 15 years in those patients with diploid cytogenetics compared with only 6 years in those who have complex abnormalities.4
Conventional cytogenetic analysis detects chromosome abnormalities in up to 50% of patients with CLL. The most frequent chromosomal changes are deletion of 13q, 11q, 17p, 6q, and trisomy 12.5 However, given the low mitotic activity of the CLL clone (most are in the G0 phase), conventional cytogenetic techniques may not detect the full complement of chromosome changes. Fluorescence in situ hybridization (FISH) can identify chromosome abnormalities in up to 80% of patients with CLL by detecting aberrations in both dividing cells and interphase nuclei.6 FISH may reveal clonal changes in some patients found to have normal karyotypes. Although cytogenetic analysis can detect the 13q deletion in 20% of cases, FISH increases the detection of 13q deletion in more than 50% of cases.7
Deletions of 11q and 17p are poor prognostic features for patients with CLL and lead to a shortened survival. Conversely, the presence of 13q deletion confers a good prognosis.7 The genetic significance of 13q deletion is not fully known. The chromosome band 13q14 carries the retinoblastoma gene (RB1), and it was originally thought that abrogation of this tumor suppressor gene was implicated in the pathogenesis of CLL. However, biallelic deletion of RB1 is rare, suggesting that there were other genes involved. One study of 75 patients with CLL showed that 33 (44%) carried either monoallelic or biallelic deletion of 13q. The deletion frequency was 16 of 75 (21%) at the RB1 gene, 33 of 75 (44%) at the D13S319 locus, and 29 of 75 (39%) at the D13S25 locus. In 16 of 75 (21%) patients, a large deletion covering all 3 of these loci was found.8 Other mechanisms may be responsible for clonality,9 including reciprocal translocations involving band 13q14 that are associated with D13S25. Another area thought to be implicated in the disease process is D13S272. However, subsequent mutation analysis of 2 candidate genes, Leu1 (deleted in lymphocytic leukemia 1) and Leu2, did not show inactivation of these genes. More recent studies show that microRNA genes miR-15a and miR-16-1 map to 13q14.3. Allelic loss in this area correlated with deletion of genes in CLL, suggesting that these 2 microRNAs can function as tumor suppressor genes.10
It has been hypothesized that homozygous deletion of the 13q locus might lead to a more accelerated disease trajectory. In this study, an analysis was performed in patients with CLL and either monoallelic or biallelic 13q deletion to detect any prognostic differences.
MATERIALS AND METHODS
Patients
We searched the electronic database for patients with CLL who carried either monoallelic or biallelic deletion of 13q and presented to the The University of Texas MD Anderson Cancer Center (MDACC) between 2002 and 2008. Any patient who carried other chromosome changes in addition to 13q and those who had received treatment prior to FISH analysis were excluded. The patients were separated into 2 groups: one group had monoallelic deletion of 13q (monoallelic group), whereas the other group had biallelic deletion of 13q (biallelic group). Any patient with 2 separate clones harboring both monoallelic and biallelic deletion of 13q was categorized as having a biallelic deletion for this analysis. A waiver of consent was obtained by the MDACC Institutional Review Board.
Treatment
Patients with CLL were treated if they had standard indications for therapy.11 The response criteria were those defined by the National Cancer Institute -sponsored working group.12 The treatment history, including time from diagnosis to first treatment and the type and duration of treatment, was recorded.
Cytogenetic and Molecular Studies
All patients except 3 had standard cytogenetic analysis with 20 cells in metaphase analyzed. All patients had FISH analysis on peripheral blood or bone marrow lymphocytes. Vysis multicolor DNA probes (Vysis-Abbott, Downers Grove, Ill) were designed to simultaneously detect the 11q22.3 (ataxia telangiectasia mutated [ATM] gene) region of chromosome 11, 17p13.1 (tumor protein p53 [TP53] gene) region of chromosome 17, the alpha satellite, centromeric region of chromosome 12 (D12Z3), the D13S319 locus (between RB1 and D13S25 loci) in the 13q14.3 region of chromosome 13, and the 13q34 region (lysosomal-associated membrane protein 1 [LAMP1] gene) near the subtelomere of chromosome 13q. A total of 200 interphases were analyzed with each probe.
Endpoints and Characteristics
Demographic information, including age, sex, and ethnicity, was reported for each patient, and the Rai stage was recorded. Pretreatment physical examination characteristics, including splenomegaly, number of lymph node sites, and presence of constitutional symptoms, were recorded. Baseline laboratory values obtained at MDACC included white blood cell count, hemoglobin level, platelet count, lactate dehydrogenase (LDH), alkaline phosphatase, albumin, electrolytes, and B2-microglobulin. Prognostic factors, including the mutation status of immunoglobulin heavy chain (VH), and the presence of both zeta-chain-associated protein kinase 70 (ZAP70) and clusters of differentiation 38 (CD38), were assessed.
Statistical Methods
For the quantitative variables, median values with range were reported, with comparison made by Mann-Whitney analysis. Chi-square analysis was performed for all categorical variables. Using Kaplan-Meier estimates, a time-to-treatment curve was generated from the date of presentation to MDACC (FISH analysis) to the time of treatment. Given the paucity of deaths in both groups, an overall survival was not estimated.
RESULTS
Patient Characteristics
FISH identified a total of 176 patients with CLL who carried 13q deletion as the sole chromosome abnormality. Of these patients, 143 had monoallelic deletion of 13q (81%) and 33 (19%) had biallelic deletion. Of the 33 patients with biallelic deletion, 18 (55%) had separate clones that had both monoallelic and biallelic 13q deletions (combined). The median time from diagnosis of CLL to presentation at MDACC was 2.1 months in the monoallelic group versus 2.2 months in the biallelic group. Quantitative variables were compared between the groups. The only 2 variables found to be significantly different between the biallelic and monoallelic group were albumin (4.5 g/dL vs 4.4 g/dL; P = .01) and ZAP70 expression (1.7% vs 4.8%; P = .010). Other values that approached significance include percent mutation of the VH gene (6% vs 4.1%; P = .06), LDH (456 IU/L vs 481 IU/L; P = .07), and age at diagnosis (60 years vs 55 years; P = .054) (Table 1).
Table 1.
Characteristics of Patients With Monoallelic Versus Biallelic Deletions of 13q
| Characteristic | Monoallelic Deletion 13q (N = 143), Median (range) | Biallelic Deletion 13q (N = 33), Median (range) | P |
|---|---|---|---|
| Diagnosis to MDACC, mo | 2.1 (0–11.7) | 2.2 (0–11.7) | .84 |
| Age at diagnosis, y | 55 (30–88) | 60 (45–75) | .054 |
| White blood cell count, × 109/L | 20 (5.1–150.5) | 18.9 (5.5–88.8) | .65 |
| Absolute lymphocytes, × 109/L | 14.9 (0–144.5) | 12.5 (3.9–79) | .789 |
| Hemoglobin, g/dL | 14.1 (7.5–17.3) | 14.3 (11.1–17.2) | .63 |
| Platelet count, × 109/L | 223 (19–554) | 232 (124–519) | .699 |
| Lactate dehydrogenase, IU/L | 481 (276–1081) | 456 (322–669) | .07 |
| Alkaline phosphatase, IU/L | 73 (15–700) | 70 (35–113) | .419 |
| Albumin, g/dL | 4.4 (3.2–5.1) | 4.5 (4–4.9) | .011 |
| B2 Microglobulin, mg/L | 2.1 (1.1–8.1) | 2.0 (1.2–4.8) | .27 |
| Immunoglobulin heavy chain, VH, %b | 95.9 (81.7–100) | 94.0 (88–100) | .058 |
| ZAP70 | 4.8 (0.4–74.4) | 1.7 (0.2–34.4) | .01 |
| CD19+CD38 | 3.65 (0–96) | 3.60 (1–94) | 1.0 |
| Sex, no. (%) | |||
| Male | 88 (62) | 18 (55) | .459 |
| Female | 55 (38) | 15 (45) | |
| Rai stage, no. (%) | |||
| 0 | 54 (38) | 12 (36) | |
| 1 | 70 (49) | 16 (48) | .478 |
| 2 | 11 (8) | 5 (15) | |
| 3 | 3 (2) | 0 (0) | |
| 4 | 5 (3) | 0 (0) | |
| Spleen size, no. (%)a | |||
| 0 cm | 131 (92) | 29 (88) | .503 |
| >0 cm | 12 (8) | 4 (12) | |
| Nodal sites involved, no. (%) | |||
| 0 | 59 (41) | 12 (36) | |
| 1 | 26 (18) | 4 (12) | .322 |
| 2 | 27 (19) | 11 (33) | |
| 3 | 31 (22) | 6 (18) | |
| ZAP70, no. (%) | |||
| Not analyzed | 38 (27) | 6 (18) | |
| Negative | 69 (48) | 17 (52) | .787 |
| Positive | 36 (25) | 10 (30) | |
Abbreviations: CD, clusters of differentiation; MDACC, The University of Texas MD Anderson Cancer Center; ZAP70, zeta-chain-associated protein kinase 70.
Spleen size measured by palpation below costal margin.
Concordance with germline.
Survival
At the time of last follow-up, none of the 33 patients (0%) in the biallelic group had died, whereas 3 of 143 patients (2%) in the monoallelic group had died. Of these 3 patients, 2 did not receive any treatment and died, respectively, of complications of metastatic colon cancer and fungal pneumonia/respiratory failure.
Time to Treatment
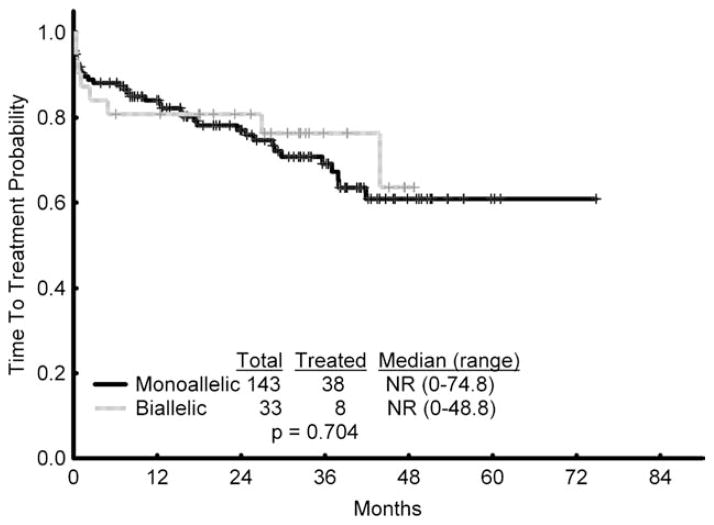
A total of 8 of 33 patients in the biallelic group (24%) and 38 of 143 (27%) patients in the monoallelic group required treatment (P = nonsignificant). The median time from presentation at MDACC to treatment was not reached in either the biallelic or monoallelic groups at the time of last follow-up (P = nonsignificant). (Fig. 1).
Figure 1.
Time to treatment from presentation to The University of Texas MD Anderson Cancer Center is shown (monoallelic vs biallelic deletion of 13q). NR indicates not reached.
Therapy and Response
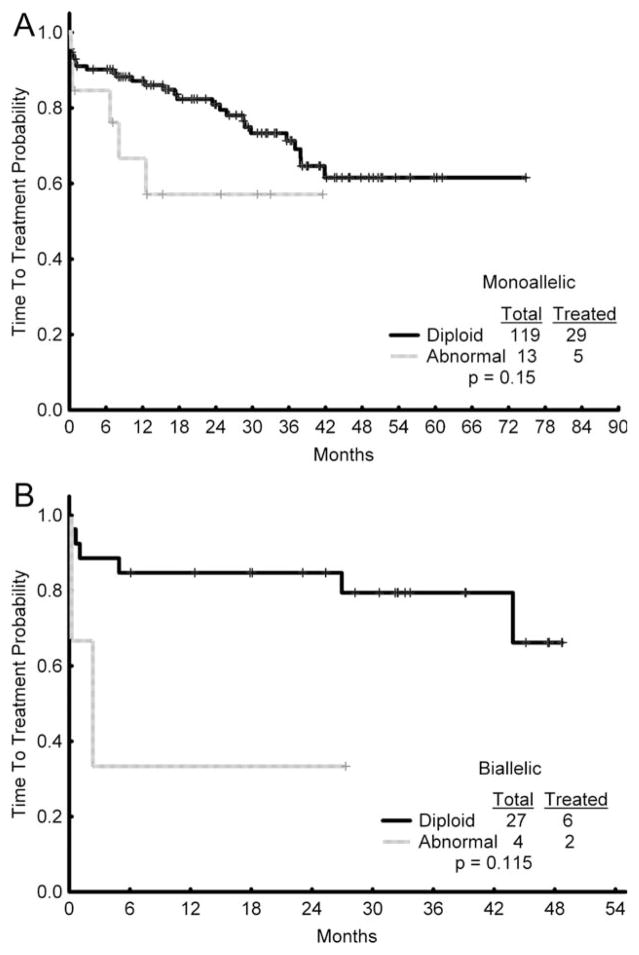
Of the 143 patients in the monoallelic group, 119 had diploid cytogenetics (83%), 13 had chromosomal abnormalities (9%), and 11 (8%) did not have a baseline bone marrow evaluation available for analysis. Of the 105 patients who were not treated, 90 had diploid cytogenetics (86%), 8 had abnormal cytogenetics (8%), and 7 (7%) did not have cytogenetics available. Of the 38 patients (27%) with monoallelic deletion who were treated, 29 (76%) had diploid cytogenetics, 4 (11%) had no baseline cytogenetics available, and 5 (13%) had abnormal cytogenetics. A total of 29 of 119 (24%) patients with diploid cytogenetics required treatment versus 5 of 13 patients (38%) with abnormal cytogenetics (P = .15) (Fig. 2A). The most common treatment regimens included fludarabine, cyclophosphamide, and rituximab (FCR) in 22 (58%) patients, cyclophosphamide, fludarabine, rituximab, and alemtuzumab (CFAR) in 4 (11%) patients, rituximab/sargramostim in 6 (16%) patients, FCR+mitoxantrone in 3 (8%) patients, and lenalidomide in 3 (8%) patients.
Figure 2.
Baseline cytogenetic status and treatment are shown in the (A) monoallelic and (B) biallelic 13q deletion groups.
Of the 33 patients in the biallelic group, 27 had diploid cytogenetics (82%), 4 had chromosomal abnormalities (12%), and 2 (6%) did not have a baseline cytogenetic analysis. Of the 8 patients with biallelic deletion who were treated (24%), 6 (75%) had diploid cytogenetics and 2 (25%) had abnormal cytogenetics (Fig. 2A). A total of 6 of 27 (22%) patients with diploid cytogenetics required treatment compared with 2 of 4 patients (50%) who had cytogenetic abnormalities (P = .115) (Fig. 2B). FCR was given to 4 patients (50%), rituximab/sargramostim was administered to 2 patients (25%), CFAR to 1 patient (12.5%), and lenalidomide to 1 patient (12.5%).
The percentage of nuclei harboring 13q deletion in both the monoallelic and biallelic treatment groups was compared. Although the median percentage of 13q deletion was higher in both the monoallelic (42.3% vs 31.5%; P = .254) and biallelic (50.5% vs 32%; P = .366) groups in patients requiring treatment, this did not reach significance. Repeat FISH analyses were performed on 12 patients at the time of progressive disease and initiation of therapy. All 12 patients showed an increase in the percentage of interphase cells carrying the 13q deletion, from a median of 35% to 79% of cells. The median time from FISH examination to treatment was 24 months. A total of 32 patients who received treatment had a repeat FISH analysis performed; however, the median time to treatment from baseline analysis in this group was only 1 month.
DISCUSSION
Our results demonstrate no significant difference in time to treatment between patients with CLL who have either monoallelic or biallelic deletion of 13q. In addition, except for the difference in ZAP70 expression and albumin levels (both of which were higher in the monoallelic group, but of no clinical significance), there was no difference in other baseline variables. Our analysis suggests that for patients with CLL who carry the 13q deletion, there is no prognostic difference between monoallelic and biallelic subgroups, and these patients could be similarly categorized.
There are several known clinical variables (ie, stage, performance status) and laboratory findings (ie, lymphocyte doubling time, LDH) that have been shown to have prognostic impact in CLL.5,13–15 In addition, markers of biologic function of the disease (ie, VH and ZAP70) can risk-stratify patients.5,16 Several studies revealed the importance of cytogenetics in risk stratification. Because FISH can examine interphase cells rather than only dividing cells, the number of cytogenetic abnormalities detected increases. However, FISH is limited to providing data on chromosomes for which specific probes are used rather than providing full chromosomal information. Deletion of 11q and 17p are associated with poor outcome in CLL; the former is highly associated with unmutated VH status and the latter with purine analogue resistance.17,18 In one study of 325 patients with CLL, the median survival time was 32 months for those with 17p deletion and 79 months for those with 11q deletion.7
Deletion of 13q is a frequent chromosomal abnormality in CLL. Two prospective multicenter studies by the German CLL Study Group (GCLLSG) found 13q deletion as the sole aberration in 44% (258 total patients) and 31% (139 total patients) of patients. When patients with 13q deletion along with additional chromosomal abnormalities were included, there was an increased number of patients on each of these studies (62% and 51%, respectively).19,20 A retrospective study that evaluated 325 patients with CLL found a sole 13q deletion in 36% of patients, and a 13q deletion coupled with other abnormalities in 55% of patients.7
Multicenter studies from the First and Second International Working Party on Chromosomes in CLL showed that 13q14 abnormalities are favorable, conferring a longer survival.4,21 Studies have shown that patients with a sole 13q deletion had the longest estimated median treatment-free interval and survival time (133 months), in comparison with those who had trisomy 12 (114 months) or normal cytogenetics (111 months).7
Several studies have demonstrated that monoallelic deletion is more frequent compared with either biallelic or combined deletions. A large study of 509 patients with CLL showed that 160 had a 13q deletion (31.4%). Of these, 122 had monoallelic deletion (76%), 18 had biallelic deletion (11.2%), and 20 had a combined deletion (12.5%). Patients with biallelic deletion of 13q were older than those with monoallelic deletion (74 years vs 68 years).22 Because this was a central laboratory–based analysis, no other clinical data were available. Another analysis of 113 patients with CLL found that 48 patients (42.5%) had deletion of 13q. Of these 48 patients, 31 had monoallelic deletion (64.5%), 9 had biallelic deletion (19%), and 8 had combined deletions (17%).23 In the current study of 176 patients with CLL and 13q deletion, 143 (81%) had monoallelic 13q deletion, 18 (10%) had combined deletion, and 15 (9%) had biallelic deletions. A preliminary study of 122 patients with 13q as the sole abnormality found similar treatment-free survival among patients with either monoallelic versus biallelic deletion of 13q.24
One study from Argentina has evaluated potential clinical differences in patients with monoallelic or biallelic deletion of 13q. A total of 103 patients were analyzed; 6 (6%) had biallelic deletion of 13q (5 with combined deletion) versus 32 (31%) with monoallelic deletion. A more rapid disease progression (6 of 6 patients [100%] vs 12 of 32 patients [38%]; P = .007), higher Rai stage (0 patients [0%] vs 15 patients [47%] with Stage 0; P = .042), higher LDH (303 IU/L vs 258 IU/L; P = .054), and more rapid time to treatment (28.5 months vs 49 months) were found in the biallelic group. No significant differences were found for age, sex, white blood cell count, hemoglobin, platelet count, and B2 microglobulin. The authors believed that biallelic deletion might portend more aggressive disease.25 Indeed, an in vitro analysis showed that patients with CLL who had biallelic deletion had more rapid lymphocyte growth kinetics compared with those in the monoallelic cohort.26 In contrast, in the present study of patients with biallelic versus monoallelic deletion of 13q, we found no significant differences in characteristics or outcome. Notably, of the 6 patients with biallelic deletion in the Argentine study, 2 were previously treated. In the present study, the analysis was performed for untreated patients seen soon after diagnosis. In addition, the current study has a substantially larger number of patients with biallelic deletion of 13q.
The prognostic impact of patients with 13q deletion could be related to its association with other biologic features. A study showed that a significant number of patients with Rai stage 0 to 1 disease had deletion of 13q (36%), suggesting that the favorable prognosis of 13q deletion could be correlated with early disease stage.27 One study demonstrated there is a significantly increased percentage of mutated immunoglobulin VH in patients with CLL who harbor either 13q deletion coupled with other chromosome abnormalities (65% vs 48%; P =.004) or with patients carrying 13q as the sole abnormality (50% vs 26%; P < .001). Conversely, there is a significantly higher percentage of unmutated immunoglobulin VH in patients carrying either 11q deletion or 17p deletions.5 Our study compared a larger number of untreated patients with both biallelic deletion (33 patients; 18 with combined deletion) and monoallelic deletion (143 patients) and did not reveal any significant difference in baseline values except for higher ZAP70 expression and higher albumin level. The age of diagnosis was higher in patients with biallelic deletion (60 years vs 55 years; P = .054) compared with those with monoallelic deletion. We found that the percentage of patients treated in the biallelic deletion group (8 of 33 [24%]) and monoallelic deletion group (38 of 143 [27%]) was identical. The median time to treatment was not significantly different. In addition, the survival rate was comparable (0 of 33 deaths [0%] in the biallelic group vs 3 of 143 patients [2%] in the monoallelic group).
To our knowledge, the largest study to date on the prognostic significance of monoallelic versus biallelic deletion of 13q was recently reported.28 The outcome of 323 treatment-naive patients with CLL who harbored isolated 13q deletion was analyzed. After a median follow-up of 2.1 years, 52 (16%) patients had been treated and 30 (9%) patients died with a median time to first treatment of 6.9 years and an overall survival (OS) of 9.3 years, respectively. There was a similar time to first treatment and OS in patients with monoallelic (n = 194), biallelic (n = 61), and mosaic (n = 68) deletion of 13q. In addition, a higher percentage of nuclei with 13q deletion (defined as > 65.5% of nuclei) was associated with significantly shortened time to first treatment (P < .001), and this remained an independent predictor after controlling for other factors, such as ZAP70, immunoglobulin VH gene rearrangement, or CD38. However, there was no association with OS. The percentage of patients remaining untreated at 5 years was 79% for patients with 13q deletion in ≤ 65.5% of nuclei versus 38% of those with 13q deletion in > 65.5% of nuclei. Our study confirmed a higher percentage of nuclei harboring the 13q deletion was present in both the monoallelic and biallelic groups for patients requiring treatment, although this finding did not reach statistical significance.
Another study confirmed the prognostic significance of the percentage of nuclei harboring 13q deletion.29 As the outcome of 109 patients with 13q deletion as the sole abnormality, patients with deletion in ≥ 80% of nuclei had a shortened time to first treatment and OS than those who had a deletion in < 80% of nuclei.
In conclusion, our study suggests that there is no prognostic difference between monoallelic versus biallelic deletion of 13q in terms of both baseline characteristics and time to treatment. With relatively short follow-up, the survival of patients in both groups is similar.
Acknowledgments
FUNDING SOURCES
No specific funding was disclosed.
Footnotes
CONFLICT OF INTEREST DISCLOSURE
The authors made no disclosure.
References
- 1.Rozman C, Montserrat E. Chronic lymphocytic leukemia. N Engl J Med. 1995;333:1052–1057. doi: 10.1056/NEJM199510193331606. [DOI] [PubMed] [Google Scholar]
- 2.Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46:219–234. doi: 10.1182/blood-2016-08-737650. [DOI] [PubMed] [Google Scholar]
- 3.Juliusson G, Merup M. Cytogenetics in chronic lymphocytic leukemia. Semin Oncol. 1998;25:19–26. [PubMed] [Google Scholar]
- 4.Juliusson G, Oscier DG, Fitchett M, et al. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323:720–724. doi: 10.1056/NEJM199009133231105. [DOI] [PubMed] [Google Scholar]
- 5.Kröber A, Seiler T, Benner A, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–1416. [PubMed] [Google Scholar]
- 6.Döhner H, Stilgenbauer S, Döhner K, Bentz M, Lichter P. Chromosome aberrations in B-cell chronic lymphocytic leukemia: reassessment based on molecular cytogenetic analysis. J Mol Med. 1999;77:266–281. doi: 10.1007/s001090050350. [DOI] [PubMed] [Google Scholar]
- 7.Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Hermanson M, Grandér D, et al. 13q Deletions in lymphoid malignancies. Blood. 1995;86:1911–1915. [PubMed] [Google Scholar]
- 9.Liu Y, Grandér D, Söderhäll S, Juliusson G, Gahrton G, Einhorn S. Retinoblastoma gene deletions in B-cell lymphocytic leukemia. Genes Chromosomes Cancer. 1992;4:250–256. doi: 10.1002/gcc.2870040310. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletion and down-regulation of micro-RNA genes miR-15 and miR-16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rai K, Patel DV. Chronic lymphocytic leukemia. In: Hoffman R, Benz EJ, Shaffil SJ, et al., editors. Hematology: Basic Principles and Practice. 3. Philadelphia, PA: Churchill Livingstone; 2000. pp. 1350–1363. [Google Scholar]
- 12.Cheson BD, Bennett JM, Grever M, et al. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood. 1996;87:4990–4997. [PubMed] [Google Scholar]
- 13.Binet JL, Auquier A, Dighiero G, et al. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer. 1981;48:198–206. doi: 10.1002/1097-0142(19810701)48:1<198::aid-cncr2820480131>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 14.Montserrat E, Sanchez-Bisono J, Viñolas N, Rozman C. Lymphocyte doubling time in chronic lymphocytic leukemia: analysis of its prognostic significance. Br J Haematol. 1986;62:567–574. doi: 10.1111/j.1365-2141.1986.tb02969.x. [DOI] [PubMed] [Google Scholar]
- 15.Di Giovanni S, Valentini G, Carducci P, Giallonardo P. Beta-2-microglobulin is a reliable tumor marker in chronic lymphocytic leukemia. Acta Haematol. 1989;81:181–185. doi: 10.1159/000205558. [DOI] [PubMed] [Google Scholar]
- 16.Crespo M, Bosch F, Villamor N, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348:1764–1775. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 17.Döhner H, Stilgenbauer S, James MR, et al. 11q deletions identify a new subset of B-cell chronic lymphocytic leukemia characterized by extensive nodal involvement and inferior prognosis. Blood. 1997;89:2516–2522. [PubMed] [Google Scholar]
- 18.Döhner H, Fischer K, Bentz M, et al. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood. 1995;85:1580–1589. [PubMed] [Google Scholar]
- 19.Bullinger L, Krautle C, Busch R, et al. Incidence and correlation of genomic aberrations with clinical and biological risk factors in B-CLL stage Binet-A within the CLL1 trial of the GCLLSG [abstract] Blood. 2001;98(suppl 1):359a. Abstract 1512. [Google Scholar]
- 20.Stilgenbauer S, Ritgen M, Bullinger L, et al. Genomic aberrations in the CLL trial of the German CLL Study Group (GCLLSG): deletion 11q23 identifies patients with molecular disease persistence after autologous high dose therapy [abstract] Blood. 2001;98(suppl 1):Abstract 3180. [Google Scholar]
- 21.Juliusson G, Oscier D, Gahrton G, et al. International Working Party on Chromosomes in CLL (IWCCLL) Cytogenetic findings and survival in B-cell chronic lymphocytic leukemia. Second IWCCLL compilation of data on 662 patients. Leuk Lymphoma. 1991;5(suppl 1):21–25. doi: 10.3109/10428199109103374. [DOI] [PubMed] [Google Scholar]
- 22.Reddy KS. Chronic lymphocytic leukaemia profiled for prognosis using a fluorescence in situ hybridization panel. Br J Hematol. 2006;132:705–722. doi: 10.1111/j.1365-2141.2005.05919.x. [DOI] [PubMed] [Google Scholar]
- 23.Dewald G, Brockman S, Paternoster S, et al. Chromosome anomalies detected by interphase fluorescence in situ, hybridization; correlation with significant biological features on B-cell chronic lymphocytic leukaemia. Br J Haematol. 2003;121:287–295. doi: 10.1046/j.1365-2141.2003.04265.x. [DOI] [PubMed] [Google Scholar]
- 24.Fink S, Geyer S, Shanafelt, et al. Clinical significance of homozygous D13S319 deletion in b-cell chronic lymphocytic leukemia (B-CLL) [abstract] Blood. 2004;2799:766a. Abstract 104. [Google Scholar]
- 25.Chena C, Avalos J, Bezares R, et al. Biallelic deletion 13q14.3 in patients with chronic lymphocytic leukemia: cytogenetic, FISH and clinical studies. Eur J Haematol. 2008;81:94–99. doi: 10.1111/j.1600-0609.2008.01086.x. [DOI] [PubMed] [Google Scholar]
- 26.Pfeifer D, Pantic M, Skatulla I, et al. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007;109:1201–1210. doi: 10.1182/blood-2006-07-034256. [DOI] [PubMed] [Google Scholar]
- 27.Ripollés L, Ortega M, Ortuño F, et al. Genetic abnormalities and clinical outcome in chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2006;171:57–64. doi: 10.1016/j.cancergencyto.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Van Dyke DL, Shanafelt TD, Call TG, et al. A comprehensive evaluation of the prognostic significance of 13q deletions in patients with B-chronic lymphocytic leukaemia. Br J Haematol. 2009;148:544–550. doi: 10.1111/j.1365-2141.2009.07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernández JA, Rodríguez AE, González M, et al. A high number of losses in 13q14 chromosome band is associated with a worse outcome and biological differences in patients with B-cell chronic lymphoid leukemia. Haematologica. 2009;94:364–371. doi: 10.3324/haematol.13862. [DOI] [PMC free article] [PubMed] [Google Scholar]