Abstract
Cardiomyocyte-like cells (CMs) derived from human pluripotent stem cells (hPSCs) present a valuable model for human disease modeling, studying early human development and, potentially, developing cell therapeutic approaches. However, the specification of early hPSC-derived CMs into defined cardiac subtypes such as atrial and ventricular cells is not well understood and, thus, poorly controlled. Moreover, the maturation status of hPSC-CMs is not well defined, yet it is known that these cells undergo at least some degree of maturation upon longer term in vitro culture. To gain insight into this process, and to assess their developmental status, we have recently generated a data set of hPSC-CMs monitoring global changes in gene expression upon long term maintenance in vitro, in comparison to human atrial and ventricular heart samples (GEO accession number GEO: GSE64189). These data present a rich resource for evaluating the maturation status of hPSC-CMs, for identifying suitable markers for subtype-specific gene expression, as well as for the generation of functional hypotheses. Here, we provide additional details and quality checks of this data set, and exemplify how it can be used to identify maturation-associated as well as cardiac subtype-specific markers.
Keywords: Human pluripotent stem cells, Cardiac differentiation, Cardiomyocytes, In-vitro maturation, Human heart
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens/hPSC line FS3F.2/Atrial and ventricular heart biopsies |
| Sex | Male |
| Sequencer or array type | HumanHT-12 v4 |
| Data format | Raw and processed |
| Experimental factors | hPSC-derived CMs: untreated cells harvested at weekly intervals |
| Experimental features | hPSCs were differentiated into cardiomyocytes and maintained under adherent culture conditions in low fetal calf serum-containing media. Samples were taken at weekly intervals up to 8 weeks. Human atrial and ventricular biopsy samples served as in vivo reference. |
| Consent | Written informed consent was obtained from patients where applicable. |
| Sample source location | Münster, Germany |
Direct link to deposited data
Deposited data can be found here: http://www.dtd.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64189.
Experimental Design, Materials and Methods
Generation and processing of samples
Cell culture. hPSCs, cell line FS3F.2 [1], were maintained in FTDA medium [2], on Matrigel™-coated dishes. Cardiac differentiation was induced as described [3]. In brief, fully confluent hPSC cultures were harvested using Accutase™ digestion, and replated onto Matrigel-coated 24-well plates (500,000 cells per well in 2 ml of day 0 differentiation medium). An aliquot of cells was used for RNA isolation (0 week time-point). Day 0 differentiation medium contained Knockout™ DMEM, insulin/transferrin/selenium, 10 μM Y27632, penicillin/streptomycin/l-Glutamine, 10 ng/ml FGF2, 1 ng/ml BMP4, and 1 μM CHIR99021 [3]. Day 1 medium contained Knockout™ DMEM transferrin/selenium penicillin/streptomycin/l-Glutamine, and 250 μM phospho-ascorbate (TS medium). On days 2 and 3, cells were fed with TS medium supplemented with 2 μM of WNT inhibitor IWP-2. Hence after, cells were maintained in basal TS medium. Spontaneous beating was observed from day 6 onwards. CM differentiation efficiency was above 85% as judged by FACS counting [3]. On day 7, the cells were harvested using 1 × TrypLE Select (Life Technologies), and pooled from independent samples. An aliquot of cells was used for RNA isolation (1 week time-point), and the remaining cells replated at ~ 250,000 cells per well of a Matrigel-coated 24-well plate, in 2% fetal calf serum/Knockout DMEM/penicillin/streptomycin/l-Glutamine. Thereafter, medium was replaced every 3–4 days. Samples of maturating hPSC-CMs were taken at weekly intervals up to 8 weeks, to be subjected to microarray analysis. Total RNA was isolated using Qiagen RNeasy columns with on-column DNA digestion.
Human heart samples. RNA samples from left and right atrial appendages have been previously described [4]. RNA samples were pooled from six independent patients per tissue type. The human left and right ventricular RNA samples were from a commercial supplier (Biocat #R1234138‐50‐BC and #R1234139‐50‐BC, respectively).
500 ng of total RNA from each biological sample was used as input for the generation of biotin-labeled cRNA using an Illumina® TotalPrep™ RNA amplification kit (Life Technologies). Following the manufacturer's instructions, in vitro transcription of double-stranded cDNA was performed for 14 h, in a PCR cycler. Purified biotin-labeled cRNA was eluted in a volume of 100 μl and quality-checked on a 2100 Bioanalyzer device (Agilent Technologies). cRNA samples were adjusted to 150 ng/μl in water, and hybridized onto Illumina HumanHT-12 v4 bead arrays following the manufacturer's instructions throughout. Hybridization was carried out at 58 °C for 18 h. Staining with streptavidin-Cy3 (GE Healthcare #PA43001) was carried out as recommended, at a concentration 1 μg/ml in blocking buffer. Dried bead arrays were scanned on a HiScan SQ device (Illumina) using default settings.
Technical and biological data quality assessment
Scanned images were confirmed to show an overall clean fluorescence spot morphology with high signal-to-noise ratio, and array data were confirmed to display an average P95 intensity of > 800 (a.u.). Inspection of raw data in GenomeStudio suggested an overall high hybridization stringency, according to internal mismatch control probes, and no major hybridization artifacts. Following these routine checks, all separately hybridized samples were background-subtracted and normalized using the Cubic Spline algorithm in GenomeStudio. This revealed a high degree of similarity between the left/right human heart samples, suggesting that they could be combined in silico. To assess overall human heart-specific gene expression regardless of chamber-specific differences, data was additionally analyzed by combining all human atrial and ventricular samples using GenomeStudio software.
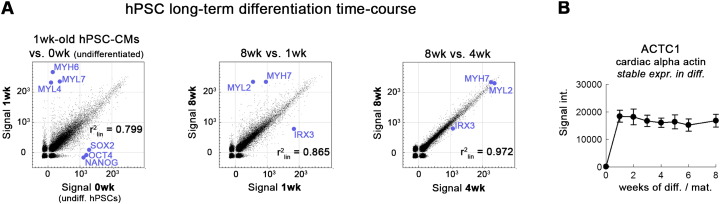
As a biological quality control step, known markers were used to assess differential gene expression between the distinct types of samples. In line with the expectations, hPSC-specific genes OCT4, NANOG and SOX2 were only expressed in the undifferentiated (0 week) cells. Conversely, structural cardiac markers (MYH6, MYL4, MYL7) were indeed only expressed in the differentiated samples and not in the undifferentiated cells (Fig. 1A, left). Focusing on gene expression changes upon long-term in vitro culture, maturation markers such as MYL2 and MYH7 were upregulated in the late (8 weeks) samples, whereas markers of immature hPSC-CMs were indeed overrepresented in the early (1 week) samples (Fig. 1A, middle). As supported by functional assays [3], however, there were only marginal differences between 4 weeks and 8 week-old hPSC-CMs, suggesting that the cells reach a rather stable transcriptomic state from approximately 4 weeks onwards (Fig. 1A, right). Furthermore, the expression pattern of the pan-cardiac marker ACTC1 (cardiac muscle alpha actin) served to indicate an overall stable cardiomyocyte signature in all differentiated samples (1 to 8 weeks, Fig. 1B).
Fig. 1.
Biological quality assessment. (A) Scatter plot analysis (power scale) of early hPSC-CMs versus undifferentiated hPSCs (left), late vs. early hPSC-CMs (middle), and 8 week vs. 4 week-old hPSC-CMs (right). Linear correlation coefficients are provided as a measure for global transcriptome similarity. Blue colored dots indicate data points of known marker genes. See text for discussion. (B) ACTC1 as a pan-cardiac marker is expressed at similar levels in all differentiated in vitro samples (from 1 week onwards). Error bars indicate bead standard deviation extracted from GenomeStudio.
Basic data analysis
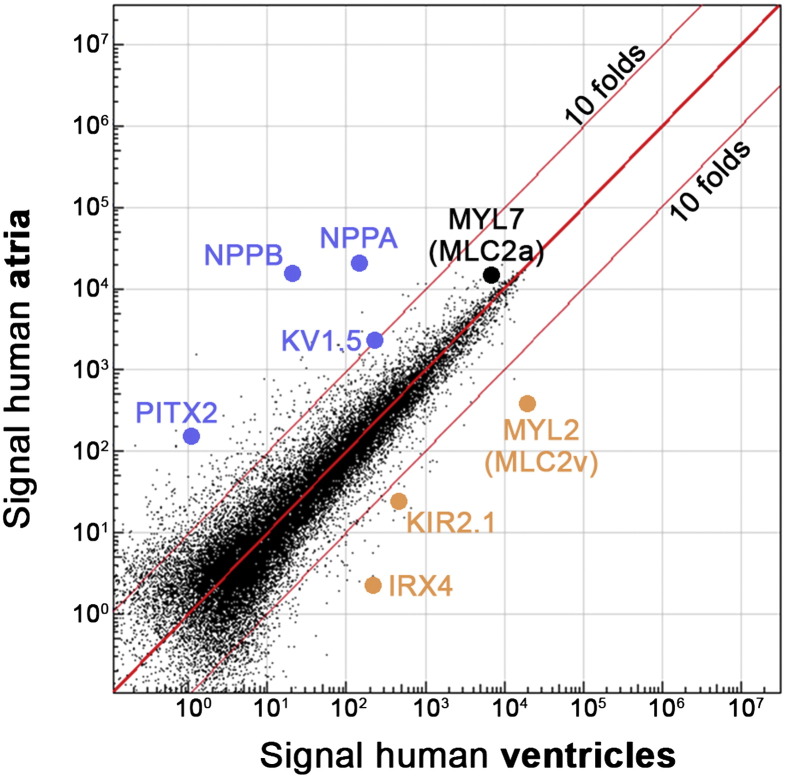
A comparison of human atrial and ventricular samples allowed for the identification of marker genes. Using stringent filtering criteria (> 10-fold differences in gene expression), these included known structural genes, ion channels, as well as transcriptional regulators (Fig. 2, Table 1). For instance, myosin light chain 2 (MYL2, also known as MLC2v) presents a rather stringent ventricular marker. By contrast, the frequently used MYL7 (MLC2a) was only about 2-fold enriched in atrial tissue, suggesting that it does not well discriminate between human cardiac subtypes. Instead, the natriuretic peptide-encoding genes NPPA and NPPB are excellent atrial markers according to this analysis.
Fig. 2.
Comparison of adult human atrial and ventricular tissue (scatter plot of combined left/right samples). Selected marker genes are highlighted by colored dots. Note that MYL7 (MLC2a) is also highly expressed in ventricular tissue.
Table 1.
Marker genes discriminating between human atrial and ventricular tissue and corresponding expression levels in late (4–8 weeks) hPSC-CMs. P values for differential gene expression are below 0.01 in all cases.
| Symbol | Atria signal | Ventricles signal | Fold change (V/A or A/V) | hPSC-CMs 4–8 weeks signal | Definition |
|---|---|---|---|---|---|
| Human ventricular markers | |||||
| DLK1 | 5 | 341 | 68 | 924 | Delta-like 1 homologue (Drosophila) |
| IRX4 | 5 | 204 | 41 | 252 | Iroquois homeobox 4 |
| MYL2 | 444 | 18,009 | 41 | 6769 | Myosin, light polypeptide 2, regulatory, cardiac, slow |
| XDH | 5 | 188 | 38 | 33 | Xanthine dehydrogenase |
| TMEM190 | 7 | 255 | 37 | 46 | Transmembrane protein 190 |
| HYAL2 | 16 | 532 | 34 | 112 | Hyaluronoglucosaminidase 2 |
| CPNE4 | 20 | 650 | 32 | Below detection | Copine IV |
| CYP1A1 | 5 | 158 | 32 | Below detection | cytochrome P450, family 1, subfamily A, Polypeptide 1 |
| IRX5 | 14 | 387 | 27 | 42 | Iroquois homeobox protein 5 |
| C3orf23 | 5 | 112 | 22 | 24 | Chromosome 3 open reading frame 23 |
| Human atrial markers | |||||
| NPPB | 15,780 | 19 | 824 | 2043 | Natriuretic peptide precursor B |
| HAMP | 1548 | 5 | 310 | 29 | Hepcidin antimicrobial peptide |
| MYBPHL | 922 | 5 | 184 | Below detection | Myosin binding protein H-like |
| NPPA | 21,892 | 143 | 153 | 1615 | Natriuretic peptide precursor A |
| PLA2G2A | 478 | 5 | 101 | Below detection | Phospholipase A2, group IIA (platelets, synovial fluid) |
| COMP | 396 | 5 | 79 | Below detection | Cartilage oligomeric matrix protein |
| TCEAL2 | 761 | 12 | 63 | 19 | Transcription elongation factor A (SII)-like 2 |
| SLPI | 274 | 5 | 55 | Below detection | Secretory leukocyte peptidase inhibitor |
| DHRS9 | 1441 | 28 | 52 | 2251 | Dehydrogenase/reductase (SDR family) member 9 |
| HP | 1316 | 27 | 49 | Below detection | Haptoglobin |
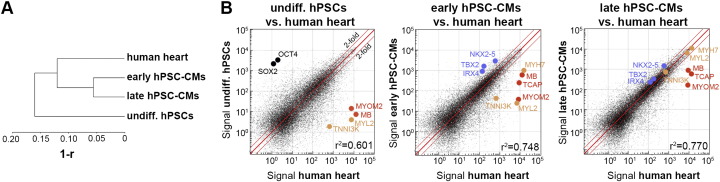
Analyzing the hPSC-based together with the in vivo data allows determination of shared and divergent gene expression between hPSC-CMs and human heart. In line with the expectations, scatter plot and clustering analyses between human heart versus (i) undifferentiated hPSCs, (ii) early (1 week) hPSC-CMs, and (iii) maturated hPSC-CMs (combined 4–8 weeks samples) suggest that over time, hPSC-CMs tend to become more similar to the human adult heart reference (Fig. 3). This tendency is also supported by the fact that a set of known immature CM markers (NKX2.5, IRX4, and others) declines upon long-term in vitro culture, whereas a set of maturation marker genes (MYH7, MYL2, and others) reached human heart-like expression levels over time (blue and orange colored genes, respectively, in Fig. 3). However, even in late hPSC-CMs a number of human heart genes were not expressed, and the global similarity to the in vivo reference appears to be rather limited (red colored genes in Fig. 3, Table 2).
Fig. 3.
Comparison of hPSC-CMs and human heart tissue. (A) Global correlation-based dendrogram showing early (1 week-old) and late (combined 4–8 weeks-old) hPSC-CMs clustering closer to human heart (combined atrial and ventricular samples) than to undifferentiated hPSCs. (B) Scatter plots comparing the indicated samples. Examples of genes enriched in the four types of samples are highlighted by different colors (black: hPSCs, blue: early hPSC-CMs, orange: late hPSC-CMs and human heart, red: human heart).
Table 2.
Selected genes enriched in early hPSC-CMs, late hPSC-CMs, and human heart. P values for differential gene expression are below 0.01 in all cases.
| Symbol | hPSCs 0 week signal | hPSC-CMs 1 week signal | hPSC-CMs 4–8 weeks signal | Human heart atr. & ventr. signal | Definition |
|---|---|---|---|---|---|
| Genes upregulated in early hPSC-CMs | |||||
| NKX2-5 | Below detection | 3201 | 1549 | 583 | NK2 transcription factor related, locus 5 |
| IRX4 | 67 | 1036 | 276 | 132 | Iroquois homeobox 4 |
| TBX2 | Below detection | 1536 | 331 | 152 | T-box 2 |
| COL2A1 | 20 | 694 | 39 | Below detection | Collagen, type II, alpha 1 |
| ISL1 | 23 | 269 | 17 | Below detection | ISL1 transcription factor, LIM/homeodomain |
| HAND1 | Below detection | 4480 | 1231 | 632 | heart and neural crest derivatives expressed 1 |
| ID2 | 110 | 1339 | 83 | 228 | Inhibitor of DNA binding 2 |
| LEF1 | 15 | 501 | 47 | Below detection | Lymphoid enhancer-binding factor 1 |
| IRS1 | 53 | 546 | 241 | 117 | Insulin receptor substrate 1 |
| MDK | 23 | 532 | 201 | Below detection | Midkine (neurite growth-promoting factor 2) |
| Genes upregulated in late hPSC-CMs and human heart | |||||
| MYH7 | Below detection | 938 | 11,781 | 13101 | Myosin, heavy chain 7, cardiac muscle, beta |
| MYL2 | Below detection | 41 | 6838 | 7737 | Myosin, light polypeptide 2, regulatory, cardiac, slow |
| TNNI3K | below detection | 46 | 757 | 659 | TNNI3 interacting kinase, transcript variant 2 |
| HSPB7 | Below detection | 3339 | 5428 | 5614 | Heat shock 27 kDa protein family, member 7 |
| PLN | Below detection | 1534 | 4645 | 4638 | Phospholamban |
| CSRP3 | below detection | 540 | 1297 | 2317 | Cysteine and glycine-rich protein 3 |
| ACTN2 | Below detection | 813 | 1130 | 1739 | Actinin, alpha 2 |
| RBM20 | below detection | 440 | 1121 | 1426 | RNA binding motif protein 20 |
| TRIM63 | Below detection | 367 | 1116 | 1544 | Tripartite motif-containing 63 |
| CORIN | Below detection | 444 | 934 | 976 | Corin, serine peptidase |
| Genes upregulated in human heart | |||||
| CASQ2 | Below detection | 10 | 23 | 6699 | Calsequestrin 2 (cardiac muscle) |
| MB | Below detection | 727 | 633 | 12450 | Myoglobin, transcript variant 1 |
| MYOM2 | 14 | 41 | 180 | 8272 | Myomesin (M-protein) 2 |
| TCAP | Below detection | 264 | 922 | 8830 | Titin-cap (telethonin) |
| MYH11 | below detection | 147 | 201 | 1835 | Myosin, heavy chain 11 |
| TNNI3 | 34 | 331 | 401 | 9157 | Troponin I type 3 (cardiac) |
| S100A1 | Below detection | 10 | 10 | 1482 | S100 calcium binding protein A1 |
| DES | Below detection | 14 | 56 | 4555 | Desmin |
| HRC | 24 | 1294 | 1105 | 4976 | Histidine rich calcium binding protein |
| MYOM1 | Below detection | 4787 | 4668 | 10215 | Myomesin 1, transcript variant |
Discussion
Despite the fact that neither the in vitro-derived nor the in vivo samples consisted of pure populations of cardiomyocytes, this data set suggests that meaningful biological information can be extracted from it. The combined hPSC-CM/adult human heart data hence presents a useful resource for evaluating the maturation status of hPSC-CMs at the transcriptional level as well as for assessing cardiac subtype-specific gene expression. Notably, according to our analysis, the frequently used MYL7 (MLC2a) gene appears to be unsuited for discriminating between atrial and ventricular subtypes. Our data instead suggests alternative genes, such as NPPA and NPPB, as being well-suited markers for evaluating atrial subtype specification in hPSC-CMs.
Acknowledgments
This work was supported by the Chemical Genomic Centre of the Max Planck Society.
References
- 1.Greber B. FGF signalling inhibits neural induction in human embryonic stem cells. EMBO J. 2011;30(24):4874–4884. doi: 10.1038/emboj.2011.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frank S., Zhang M., Scholer H.R., Greber B. Small molecule-assisted, line-independent maintenance of human pluripotent stem cells in defined conditions. PLoS One. 2012;7(7):e41958. doi: 10.1371/journal.pone.0041958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang M. Universal cardiac induction of human pluripotent stem cells in 2D and 3D formats — implications for in-vitro maturation. Stem Cells. 2015 doi: 10.1002/stem.1964. [DOI] [PubMed] [Google Scholar]
- 4.Kahr P.C. Systematic analysis of gene expression differences between left and right atria in different mouse strains and in human atrial tissue. PLoS One. 2011;6(10):e26389. doi: 10.1371/journal.pone.0026389. [DOI] [PMC free article] [PubMed] [Google Scholar]