Abstract
Macrophages are important for mounting inflammatory responses to tissue damage or infection by invading pathogens, and therefore modulation of their cellular functions is essential for the success of the immune system as well as for maintaining tissue homeostasis. Small non-coding RNAs are important regulatory elements of gene expression and microRNAs are the most widely known to be fundamental for the proper development of cells of the immune system. Macrophages can exhibit different phenotypes, depending on the cytokine environment they encounter in the affected tissues. We have analyzed the microRNA expression profiles during maturation of human primary monocytes into macrophages and polarization by pro- or anti-inflammatory cytokines. Here we describe the analysis of next-generation sequencing data deposited in EMBL–EBI ArrayExpress under accession number E-MTAB-1969 and associated with the study published by Cobos Jiménez and collaborators in Physiological Genomics in 2014 (1). The data presented here contributes to our understanding of microRNA expression profiles in human monocytes and macrophages and will also serve as a resource for novel microRNAs and other small RNA species expressed in these cells.
Keywords: microRNAs, Macrophages, Cytokines, Next-generation sequencing, DESeq
| Specifications | |
|---|---|
| Organism Cell type |
Homo sapiens Primary monocytes, monocyte-derived macrophages |
| Sequencer or array type | SOLiD 4 |
| Data format | Raw and normalized |
| Experimental factors | Cytokine treatment (IFNγ + TNFα, IL-4, IL-10) |
| Experimental features | microRNA expression in freshly isolated monocytes, compared to 5-day macrophages or cytokine treated macrophages (M1, M2a or M2c) |
| Consent | Written informed consents were obtained from all donors in accordance with the ethical principles set out in the declaration of Helsinki. This study was approved by the Medical Ethics Committee of the Academic Medical Center and the Ethics Advisory Body of the Sanquin Blood Supply Foundation in Amsterdam, The Netherlands. |
| Sample source location | Amsterdam, The Netherlands |
Direct link to deposited data
Deposited data can be found here: http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-1969/
Experimental design, materials and methods
Monocytes are derived from common myeloid progenitors and circulate in the blood stream. Once they migrate into the tissues, monocytes start differentiating into macrophages and, depending on the cytokine environment that they encounter, they can become polarized into M1 macrophages (IFNγ + TNFα/LPS), M2a macrophages (IL-4) or M2c macrophages (IL-10) [2], [3]. Previously we have shown that the incubation of freshly isolated monocytes under cytokine stimulation for 5 days allows macrophages to differentiate into cells with polarized phenotypes, which was demonstrated by the expression of characteristic surface markers [4]. Therefore we isolated human primary monocytes and allowed them to differentiate in vitro for 5 days, in the presence or absence of cytokine stimulation. Subsequently, RNA was isolated to prepare next generation sequencing libraries.
Isolation of monocytes and cell culture
Monocytes were obtained from buffy coats from 4 healthy blood donors. Written informed consents were obtained from all donors in accordance with the ethical principles set out in the declaration of Helsinki. This study was approved by the Medical Ethics Committee of the Academic Medical Center and the Ethics Advisory Body of the Sanquin Blood Supply Foundation in Amsterdam, The Netherlands. Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats using Lymphoprep (Axis-Shield, Oslo, Norway) density gradient centrifugation. Monocytes were isolated by adherence to plastic and cultured in Iscove's modified Dulbecco's medium (IMDM; Lonza, Basel, Switzerland) supplemented with 10% [v/v] heat-inactivated human pooled serum (HPS), penicillin (100 U/ml; Invitrogen, Carlsbad, CA), streptomycin (100 μg/ml; Invitrogen) and ciproxin (5 μg/ml; Bayer, Leverkusen, Germany) for 5 days in the presence of different cytokines: IFN-γ at 50 U/ml (Sigma-Aldrich), in combination with TNF-α at 12.5 ng/ml (Peprotech, Rocky Hill, NJ, USA), IL-4 at 50 ng/ml (Peprotech), IL-10 at 50 ng/ml (Peprotech) or medium alone at 37 °C in a humidified atmosphere supplemented with 5% CO2.
Library preparation and SOLiD sequencing
Total RNA was isolated from monocytes and 5-day cultured macrophages (unstimulated or cytokine-polarized), using TriPure Isolation Reagent (Roche) according to the manufacturer's instructions. RNA quality was determined using NanoDrop® Spectrophotometer (ThermoFisher Scientific, Waltham, Massachusetts, USA) and the Agilent 2100 Bioanalyzer with the RNA6000 Nano kit and the Small RNA Chip Kit (Agilent, Santa Clara, CA, USA). All samples used subsequently for sequencing had an A280/260 value higher than 1.8 and a RNA integrity number (RIN) higher than 6. Library preparation was carried out following the SOLiD™ Total RNA-Seq Kit Protocol (PN 4445374, Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol for Small RNA library preparation. Briefly, RNA samples were first enriched for their small RNA fraction, by size selection between 15 nt and 40 nt on 15% TBE-Urea acrylamide gels. The small RNA-enriched samples were hybridized and ligated for cDNA synthesis. cDNA libraries were further purified with the MinElute® PCR Purification Kit (Qiagen, Venlo, The Netherlands). After a size-selection step on 10% TBE-Urea gels, cDNA samples were amplified on gel using 19 cycles instead of 15 recommended by the manufacturer. For multiplexing purposes, the cDNA samples were amplified using the SOLiD™ 3′ Primers from the SOLiD RNA Barcoding Kit. cDNA libraries were purified using the PureLink™ PCR Micro Kit (Invitrogen, Carlsbad, CA, USA) and subjected to a second round of size selection on gel and purification according to the manufacturer's instructions. The quality of the cDNA library was assessed with a smear analysis using the Bioanalyzer 2100 software (Agilent).
cDNA libraries were pooled together at an equimolar ratio and used for the emulsion PCR reaction. Workflow Analysis (WFA) Runs showed that the bead preparation had titration metrics above 74% and Noise-to-Signal ratio below 5%. The libraries were sequenced to 35 bp read length in a 1-well deposition chamber using the Multiplex Fragment Sequencing reagents and the SOLiD™ 4 Analyzer.
Analysis of raw sequencing data
Quality control
The raw data quality was analyzed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc), which determines various sequence characteristics to identify biases in the data. First, the raw sequence color space data was converted to Fastq format, which comprises both the nucleotide sequence and the corresponding nucleotide quality scores. Subsequently, FastQC was used to determine the per base sequence quality (average quality at each nucleotide position in the reads) and the per base sequence content (proportion of each base at each nucleotide position for all reads; should be around 0.25). These quality measures were helpful to decide which samples to omit from further analysis. Several samples had a significantly lower quality at specific nucleotide positions compared to the rest of the positions in the reads. These samples also had an unbalanced base content at many positions, up to 80% per base. We continued with 14 from the 24 samples which varied in size from 1,400,000 to 8,500,000 reads.
Trimming and mapping
To quantify all expressed microRNAs in the samples, we mapped the reads directly against all mature microRNAs present in mirBase version 18 [5], which contained 1919 human microRNAs. We used BWA [6] for the mapping and made a reference index based on the fasta file containing all mature microRNAs, from which we selected the human microRNAs. Subsequently, we replaced all uracils with thymines. Prior to mapping our sequence data, we removed the remaining parts of the P2 sequencing adapter in the reads with the use of FAR (Flexible Adapter Remover, an unpublished legacy software tool based on the Needleman Wunsch alignment algorithm [7]. This resulted in libraries with reads of unequal lengths varying from 16 to 34 bases. For this trimming we required a minimum overlap of 8 bases between the read and the P2 sequence, and a minimal read length of 15 bases. During alignment we allowed for one mismatch and no gaps. After removing reads mapping to multiple microRNA and reads with a mapping quality of zero, each sample contained around 12% uniquely mapping reads which were used to produce an expression count for each microRNA. Aligned reads were counted in R using the packages Rsamtools [8] and ShortRead [9]. The counts served as input for the statistical analysis.
Statistical analysis
To identify significantly expressed microRNAs between different stimulated cells compared with unstimulated macrophages or monocytes, all pairwise comparisons were analyzed using DESeq [10], a Bioconductor [11] package which uses a negative binomial distribution for statistical testing. We let DESeq normalize the counts based on estimated library sizes. Counts from technical replicates (2 sequencing runs) were summed. We used samples with high quality and which were all isolated from one individual. DESeq is capable of statistical testing without replicates. The method is based on the assumption that the mean is a good predictor for the dispersion. Given two samples from different conditions, it is assumed that the majority of genes are not influenced by the condition and the estimated dispersion between conditions is used for the variance between replicates [10]. We also compared IFNγ + TNFα, IL-4 and IL-10 versus D0 and medium. For further analysis we plotted the log2 of the fold change (M) versus the average expression (A) and a clustering of the top 100 most varying microRNAs.
Selection and characterization of miRNAs
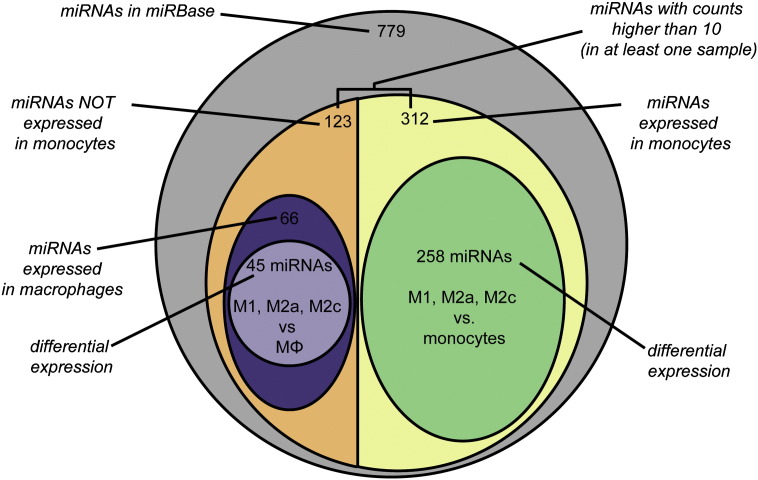
From the 1919 human miRNAs reported in miRBase, we identified 779 miRNA sequences in our samples. In order to identify miRNAs that were truly differentially regulated among monocytes and polarized macrophages, we first selected those miRNAs that had normalized counts higher than 10 in at least one of the samples analyzed (monocytes, macrophages, M1, M2a or M2c) and resulted in 435 miRNAs. These miRNAs were divided into two categories, depending on their expression in monocytes; either higher than 10 normalized counts or lower. In the first category, 123 miRNAs had normalized count values below 10 and 312 miRNAs had normalized count values higher than 10 (Fig. 1). The first category would include a group of miRNAs that is absent in monocytes but may have high expression levels in macrophages. Therefore in this category, miRNAs were selected if they expressed normalized count values higher than 10 in unstimulated macrophages (66 miRNAs, in purple, Fig. 1). From these, miRNAs were only selected if changes in expression values after comparing unstimulated cells (MΦ) with each polarized phenotype (M1, M2a or M2c) were larger than 2 times (45 miRNAs) with an absolute difference in counts of more than 10.
Fig. 1.
Selection of miRNAs. Schematic representation of the selection criteria used in this study to select miRNAs for further confirmation of their expression levels in human macrophages. The selection criteria are indicated in italic font, and the number of miRNAs selected with each criterion is indicated with a line. miRNAs were considered expressed in a cell type when they had counts higher than 10, and differentially expressed when their fold change was higher than 2, with an absolute difference higher than 10 counts.
The second category includes miRNAs that are highly regulated during maturation of monocytes into macrophages, and therefore miRNAs were selected if they showed changes in expression larger than 2 times after comparing monocytes with unstimulated macrophages (MΦ) or polarized macrophages (M1, M2a or M2c), with an absolute difference in counts of more than 10. These reduced the number of miRNAs in this category from 312 to 258 miRNAs (in green, Fig. 1). In this group there were 154 miRNAs that increased their expression levels during maturation from monocytes, and 104 miRNAs whose expression decreased during maturation.
From the two categories described above, a total of 50 microRNAs were selected for further validation. The selected miRNAs had normalized count values that represented the wide range of miRNA expression of the entire dataset, i.e. some miRNAs displayed expression values of less than 100, whereas some others had normalized count values of more than one million. Several of these microRNAs were selected because they have been described to be involved in maturation of monocytes, regulation of inflammatory responses in macrophages or to be expressed upon cytokine polarization of macrophages. Furthermore, other miRNAs were manually selected because they displayed large variation in the normalized count values among monocytes and different polarized macrophages, or because changes in expression when comparing the different polarization conditions to monocytes or unstimulated macrophages, were larger than 4 times. The expression values, fold change and absolute change in expression and selection based on reports from previous literature, are shown in Supplementary table 1.
Discussion
In this study we have collected and described a unique dataset that contains expression profiles of small RNAs in human primary monocytes and macrophages. The initial analysis of this dataset resulted in a comprehensive characterization of miRNA expression and it's relation with monocyte maturation and macrophage polarization. This data has revealed the tight relationship between miRNA expression and control of innate immune responses by macrophages [1]. Further in-depth analysis of this data set will reveal the expression of other small RNA species and will also allow for the discovery of new small RNAs in human monocytes and macrophages, and therefore the characterization of novel regulatory elements in these cells.
The following is the supplementary data related to this article.
Selection of miRNAs based on expression levels in human monocytes and macrophages.
Acknowledgments
This work was supported by The Academic Medical Center of the University of Amsterdam.
References
- 1.Cobos Jiménez V., Bradley E.J., Willemsen A.M., van Kampen A.H., Baas F., Kootstra N.A. Next-generation sequencing of microRNAs uncovers expression signatures in polarized macrophages. Physiol. Genomics. 2014;46:91–103. doi: 10.1152/physiolgenomics.00140.2013. [DOI] [PubMed] [Google Scholar]
- 2.Martinez F.O., Helming L., Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 3.Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cobos Jiménez V., Booiman T., de Taeye S.W., van Dort K.A., Rits M.A., Hamann J., Kootstra N.A. Differential expression of HIV-1 interfering factors in monocyte-derived macrophages stimulated with polarizing cytokines or interferons. Sci. Rep. 2012;2:763. doi: 10.1038/srep00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 8.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome S., Project Data Processing The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan M., Anders S., Lawrence M., Aboyoun P., Pages H., Gentleman R. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A.J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J.Y., Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Selection of miRNAs based on expression levels in human monocytes and macrophages.