Abstract
Transcriptomes are dynamic and unique, with each cell type/tissue, developmental stage and species expressing a different repertoire of RNA transcripts. Most mRNAs and well-characterized long noncoding RNAs are shaped with a 5′ cap and 3′ poly(A) tail, thus conventional transcriptome analyses typically start with the enrichment of poly(A)+ RNAs by oligo(dT) selection, followed by deep sequencing approaches. However, accumulated lines of evidence suggest that many RNA transcripts are processed by alternative mechanisms without 3′ poly(A) tails and, therefore, fail to be enriched by oligo(dT) purification and are absent following deep sequencing analyses. We have described an enrichment strategy to purify non-polyadenylated (poly(A)−/ribo−) RNAs from human total RNAs by removal of both poly(A)+ RNA transcripts and ribosomal RNAs, which led to the identification of many novel RNA transcripts with non-canonical 3′ ends in human. Here, we describe the application of non-polyadenylated RNA-sequencing in rhesus monkey and mouse cell lines/tissue, and further profile the transcription of non-polyadenylated RNAs across species, providing new resources for non-polyadenylated RNA identification and comparison across species.
Keywords: Non-polyadenylated RNAs, RNA-seq, lncRNAs, sno-lncRNAs, Species-specific
| Specifications | |
|---|---|
| Organism/cell line/tissue | Macaca mulatta and Mus musculus |
| Sex | Cell lines and Mus musculus tissue |
| Sequencer or array type | Illumina HiSeq 2000 |
| Data format | Raw data: TXT files; analyzed data: bigwig files |
| Experimental factors | Embryonic stem cell lines and Mus musculus hippocampus tissue |
| Experimental features | Non-polyadenylated (poly(A)−/ribo−) RNAs were enriched from total RNAs by removal of poly(A)+ RNA transcripts and ribosomal RNAs. Polyadenylated (poly(A)−/ribo−) RNAs were enriched from total RNAs with oligo(dT) selection. Gene expression was compared from either polyadenylated or non-polyadenylated RNA-seq. |
| Consent | Cell lines and animal tissue only |
| Sample source location | Shanghai, China |
Direct link to deposited data
Deposited data can be found at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53942.
Experimental design, materials and methods
Cell culture, RNA isolation, poly(A)−/ribo− fractionation and RNA-seq
Non-polyadenylated (poly(A)−/ribo−) RNA sequencing has been successfully performed to explore the repertoire of RNA molecules without 3′ poly(A) tails in human cell lines [1], followed by identification of new types of long noncoding RNAs (lncRNAs) in human [2], [3], [4]. However, the landscape of the non-polyadenylated RNA fraction in other species has not been documented yet.
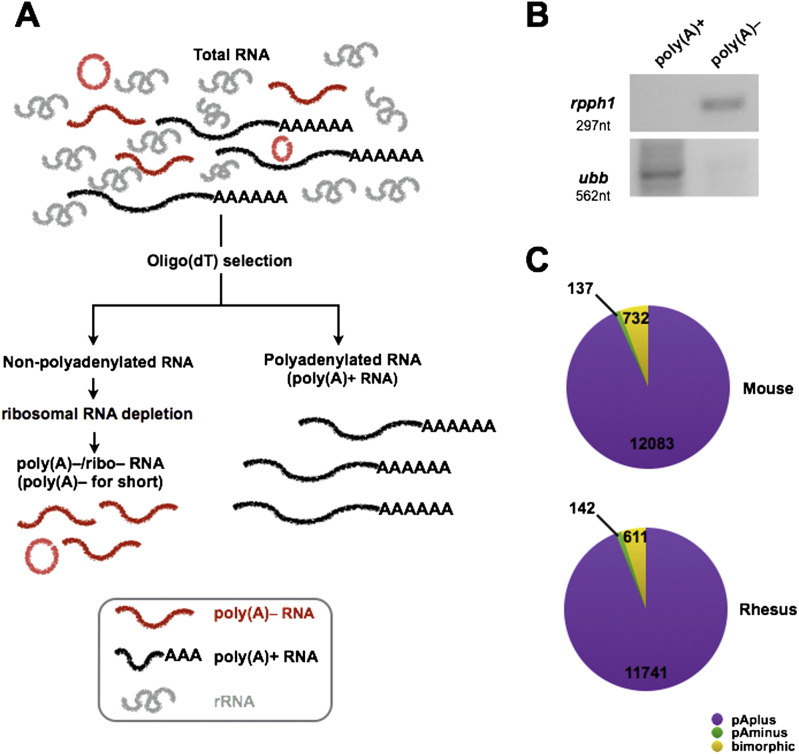
Here, we characterized non-polyadenylated RNA transcripts from two model organisms, rhesus monkey and mouse, with a similar strategy as described previously [1]. As indicated in Fig. 1A, total RNAs from R1 mouse embryonic stem cells (mESCs) and IVF3.2 rhesus monkey ESCs [5] were individually extracted with Trizol reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocols, followed by DNase I treatment (Ambion, DNA-free™ Kit) at 37 °C for 30 min to remove genomic DNA contamination. Total RNAs were then incubated with oligo(dT) magnetic beads to isolate either polyadenylated (poly(A)+) RNAs, which were bound to oligo(dT) beads, or non-polyadenylated RNAs, which were present in the flow through after incubation. Selection with oligo(dT) magnetic beads was performed three times to ensure pure poly(A)+ and non-polyadenylated RNA populations. The non-polyadenylated RNA population was further processed twice with the RiboMinus kit (Human/Mouse Module, Invitrogen, Carlsbad, CA, USA) to deplete most of the abundant ribosomal RNAs and obtain the ribosomal free non-polyadenylated (poly(A)−/ribo−) RNA population. Two representative genes, RPPH1 without a poly(A) tail (non-polyadenylated) or UBB with a poly(A) tail (polyadenylated), were examined by RT-PCR (Fig. 1B) to confirm the successful fractionation of poly(A)+ transcripts and poly(A)−/ribo− transcripts, respectively. RNA fragmentation, random hexamer-primed cDNA synthesis, linker ligation, size selection and PCR amplification were performed individually for each sample according to Illumina protocols. ~ 30 million 1 × 100 reads for each sample were acquired with the Illumina Hiseq 2000 system. Quality control checks of raw sequencing data were performed with FastQC.
Fig. 1.
(A) A schematic diagram showing the pipeline of non-polyadenylated (poly(A)−/ribo−) RNA sequencing. (B) Validation of RPPH1 and UBB in R1 mouse embryonic stem cells (mESCs) by RT-PCR. (C) Classification of poly(A)+, poly(A)− and bimorphic predominant transcripts in mouse and rhesus monkey.
RNA-seq alignment
Recently, accumulated lines of evidence have shown that non-polyadenylated RNAs are ubiquitously transcribed in the human genome [1]. To obtain comprehensive non-polyadenylated RNA alignments, sequencing reads were mapped against relevant genomes (Rhesus: rheMac3, BGI CR_1.0; Mouse: mm9, NCBI37) using TopHat 2.0.8 (parameters: −g 1 −a 6 −i 50 −microexon-search −coverage-search −m 2) with existing annotations (Rhesus: RefSeq Genes, updated on 2013/3/24; Mouse: UCSC Genes, updated on 2011/5/30). For visualization, bigWig files were generated using UCSC bedGraphToBigWig V4 from bedGraph files converted from mapped BAM files through genomeCoverageBed v2.17.0, then uploaded to the UCSC genome browser. Because gene annotations in rhesus were not complete, coordinates from human UCSC Genes annotations (knownGene.txt, updated on 2013/06/30) were converted to the rheMac3 assembly with LiftOver (parameters: −minMatch=0.1 −minBlocks=0.5 −fudgeThick) and used as rhesus gene annotations in the following analyses. Normalized gene expression levels (Reads Per Kilobase per Million mapped reads, RPKM) were calculated for all the existing genes (Rhesus: converted human gene annotations; Mouse: UCSC Genes, updated on 2011/5/30) in each sample.
Gene classification
Genes were classified into the poly(A)− predominant subgroup, the poly(A)+ predominant subgroup and the bimorphic subgroup according to their 3′ end structures using several parameters, including RPKM values for expression level, fold changes of poly(A)−/ribo− reads verse poly(A)+ reads, and P-value of fold change determined by Wald test [1]. Genes with a low expression (RPKM value < 1 in both the poly(A)−/ribo− dataset and the poly(A)+ dataset) and/or a low significant change (P-value of fold change > 0.05, Wald score > −1.96 and < 1.96) were discarded before classification.
-
1)
For genes in the poly(A)− predominant subgroup, the RPKM value from the poly(A)−/ribo− sample must be greater than or equal to 1, the fold change of the RPKM value of poly(A)−/ribo− versus the RPKM value of poly(A)+ must be greater than or equal to 2, and the P-value of fold change must be smaller than 0.05 (Wald score > 1.96).
-
2)
For genes in the poly(A)+ predominant subgroup, the RPKM value from the poly(A)+ sample must be greater than or equal to 1, the fold change of the RPKM value of poly(A)−/ribo− versus the RPKM value of poly(A)+ must be less than or equal to 0.5, and the P-value of fold change must be smaller than 0.05 (Wald score < −1.96).
-
3)
For genes in the bimorphic subgroup, the RPKM value from the poly(A)+ sample or poly(A)−/ribo− sample must be greater than or equal to 1, the fold change of the RPKM value of poly(A)−/ribo− versus the RPKM value of poly(A)+ must be between 0.5 and 2, and the P-value of fold change must be smaller than 0.05 (Wald score > 1.96 or <−1.96).
Although most RefGenes are polyadenylated in rhesus (94%) and mouse (93%), many RefGenes were also grouped into the poly(A)− predominant or the bimorphic subgroups with high expression (Fig. 1C). Of note, numerous non-RefGene transcripts were also predicted by de novo Cufflinks assembly, and their detailed classification requires further examination (data not shown).
Non-polyadenylated transcript characterization
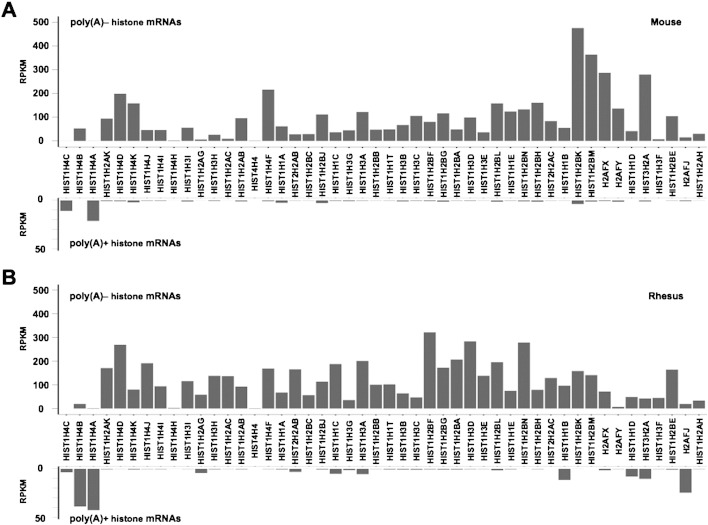
As expected [1], [6], replication-dependent histone mRNAs are mostly expressed without 3′ poly(A) tails in mouse (Fig. 2A) and rhesus (Fig. 2B) poly(A)−/ribo− RNA-seq datasets, further indicating that both fractionation and criteria for gene classification in this study are reliable. To fully characterize the constitution of the non-polyadenylated RNA fraction in rhesus and mouse, the poly(A)− predominant subgroup was further cataloged into different RNA families.
Fig. 2.
The relative expression (normalized read densities) of all histone genes in both the poly(A)−/ribo− RNA-seq dataset and the poly(A)+ RNA-seq dataset in mouse (A) and rhesus monkey (B).
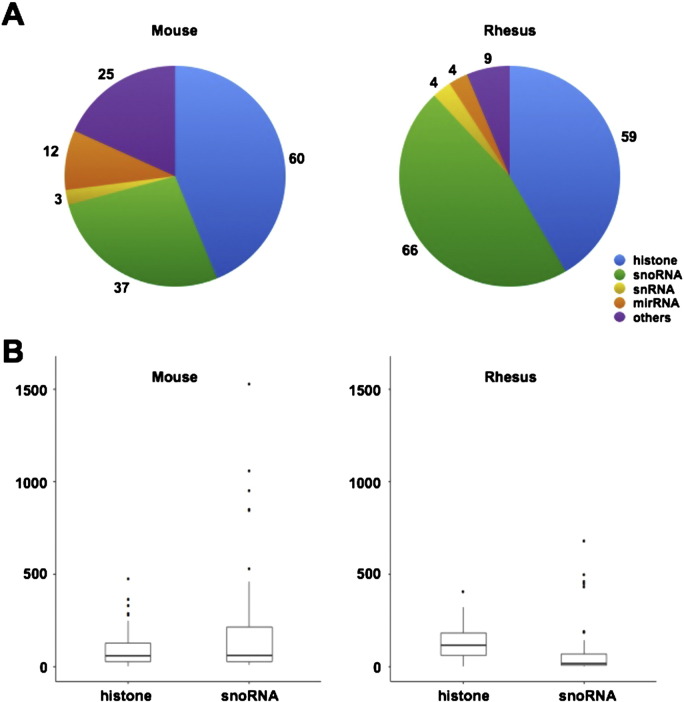
Surprisingly, a large amount of snoRNAs were found in the poly(A)− predominant subgroup of rhesus (Fig. 3A, right panel) compared with human [1] and mouse (Fig. 3A, left panel). SnoRNAs, a class of small noncoding RNAs, could generally be grouped into C/D box snoRNAs, carrying conserved boxes C (RUGAUGA, R = purine) and D (CUGA) near their 5′ and 3′ ends, and H/ACA box snoRNAs, containing the H box (ANANNA) and ACA box sequences, thus sharing high similar sequence structures that are easy to lead to false annotations. Because rhesus gene annotations were converted from human genes according to the sequence similarity, some converted snoRNA annotations in rhesus may be redundant, albeit with highly consensus sequences with human snoRNA homologs. To confirm our hypothesis, we checked the expression level of snoRNAs and histone genes in mouse and rhesus (Fig. 3B). While in mouse, snoRNAs and histone genes share a similar expression pattern, the overall expression levels of snoRNAs is low compared with histone genes in rhesus, suggesting that some lowly expressed non-snoRNA signals may obstruct the correct transcriptome profiling of snoRNAs in rhesus so as to result in many false positive snoRNAs infiltrating the poly(A)− predominant subgroup. Therefore, more biological experiments and computational predictions are required to improve gene annotations in rhesus for a better profiling of non-polyadenylated transcripts.
Fig. 3.
(A) Classification of poly(A)− transcripts in mouse (left panel) and rhesus monkey (right panel). (B) The relative expression (normalized read densities) of all histone genes and snoRNAs in the poly(A)−/ribo− RNA-seq dataset in mouse (left panel) and rhesus monkey (right panel).
Discussion
It has been reported that in the human genome lots of non-polyadenylated transcripts are novel lncRNAs with special structures, and some lncRNAs could participate in multiple layers of biological processes (such as alternative splicing [2], transcription regulation [4] and mircroRNA regulation [7], [8]). However, they (like sno-lncRNAs) showed even less conservation than other non-conserved polyadenylated lncRNAs [9], thus challenging current computational and experimental techniques. For instance, precise prediction of circular RNAs is usually a bottleneck during conventional transcriptome profiling, because of their unique circular structures [3], [4], [7]. This difficulty thus limits the subsequent analyses of their biogenesis and biological function. As a result, transcriptome profiling of non-polyadenylated RNA transcripts, a great portion of which consist of uncharacterized and lowly conserved long noncoding RNAs, is a difficult and time-consuming process, requiring deliberate and careful design for both the computational prediction pipeline and further experimental validation. However, as shown here, polyadenylated and non-polyadenylated RNA-seq datasets for rhesus and mouse, with relatively pure fractions and millions of high-quality sequencing reads, offer a particularly useful starting point for comprehensively studying the landscape of non-polyadenylated transcripts, which will aid in deciphering the molecular mechanisms driving transcriptome complexity across different species.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
We thank Dr. Christopher D. Green for proof reading, and lab members for helpful discussion and technical support. H9 cells were obtained from the WiCell Research Institute. RNA-seq was performed at CAS-MPG Partner Institute for Computational Biology Omics Core, Shanghai, China. This work was supported by grants 2014CB964800 and 2014CB910600 from MOST, 2012OHTP08 from CAS, and 31271390 from NSFC.
References
- 1.Yang L., Duff M.O., Graveley B.R., Carmichael G.G., Chen L.L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin Q.F., Yang L., Zhang Y., Xiang J.F., Wu Y.W., Carmichael G.G., Chen L.L. Long noncoding RNAs with snoRNA ends. Mol. Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 3.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 5.Sun Z., Wei Q., Zhang Y., He X., Ji W., Su B. MicroRNA profiling of rhesus macaque embryonic stem cells. BMC Genomics. 2011;12:276. doi: 10.1186/1471-2164-12-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marzluff W.F., Wagner E.J., Duronio R.J. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 8.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X.O., Yin Q.F., Wang H.B., Zhang Y., Chen T., Zheng P., Lu X., Chen L.L., Yang L. Species-specific alternative splicing leads to unique expression of sno-lncRNAs. BMC Genomics. 2014;15:287. doi: 10.1186/1471-2164-15-287. [DOI] [PMC free article] [PubMed] [Google Scholar]