Abstract
Neonatal infection remains a primary cause of infant morbidity and mortality worldwide and yet our understanding of how human neonates respond to infection remains incomplete. Changes in host gene expression in response to infection may occur in any part of the body, with the continuous interaction between blood and tissues allowing blood cells to act as biosensors for the changes. In this study we have used whole blood transcriptome profiling to systematically identify signatures and the pathway biology underlying the pathogenesis of neonatal infection. Blood samples were collected from neonates at the first clinical signs of suspected sepsis alongside age matched healthy control subjects. Here we report a detailed description of the study design, including clinical data collected, experimental methods used and data analysis workflows and which correspond with data in Gene Expression Omnibus (GEO) data sets (GSE25504). Our data set has allowed identification of a patient invariant 52-gene classifier that predicts bacterial infection with high accuracy and lays the foundation for advancing diagnostic, prognostic and therapeutic strategies for neonatal sepsis.
Keywords: Neonatal sepsis, Whole blood, Gene expression profiling, Microarray
Introduction
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens/whole blood |
| Sex | Male and female |
| Sequencer or array type | Illumina HT-12V3.0 Whole Human Genome microarray, CodeLink 55K Whole Human Genome microarray, Affymetrix U219 Whole Human Genome microarray and Affymetrix HG U133 Plus 2.0 Whole Human Genome microarray |
| Data format | Raw data (Tab delimited text files of background subtracted signals and .CEL files) |
| Experimental factors | Blood culture or cerebrospinal fluid positive bacterial sepsis vs. healthy control whole blood samples and culture negative suspected infected samples |
| Experimental features | A case–control gene expression profiling study of whole blood taken from neonates at the first clinical sign of sepsis and control healthy neonates. Study includes training and replication sets for blood culture positive samples and clinical evaluation set of blood culture negative sepsis cases. Results compared blood culture or cerebrospinal fluid positive septic neonates, blood culture negative septic neonates and healthy control neonates. Prior power calculations were based on Healthy Edinburgh neonates using the CodeLink platform and Gambian infants (9 months of age) were used for further refinement of power calculations using Illumina HT-12 platform. |
| Consent | Written informed consent was obtained from parents of all enrolled infants in accordance with approval granted by the Lothian Research Ethics Committee for blood samples for RNA isolation obtained at the first time of clinical signs of suspected sepsis (reference 05/s1103/3). Samples obtained from The Gambia conformed to MRC policy regarding ethical research in children and were approved by the local scientific coordinating committee (SCC), the Joint Gambia Government/MRC Ethics Committee and by the London School of Hygiene and Tropical Medicine Ethics Committee (reference SCC1085 Pilot Study 1 (L2008.63)) |
| Sample source location | Edinburgh, UK and The Gambia |
Direct link to deposited data
Deposited data are available here: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE25504.
Experimental design, materials and methods
Patient demographics and experimental design
The study was conducted in the Neonatal Unit, Royal Infirmary of Edinburgh and the Division of Pathway Medicine, University of Edinburgh. The patient demographics, microbial organisms isolated and reasons for blood sampling in controls for all patient sets are shown in Table 1. Infants having blood cultures taken to investigate suspected infection (Table 1B) and “well” control infants having blood taken for other clinical reasons (Table 1C) were studied. Samples taken from patients with suspected clinical infection that proved to have microbiological evidence of infection from a usually sterile body site were identified and formed the infected group. Full clinical assessment for early and late symptoms and signs of sepsis followed criteria for neonatal sepsis taken from data as detailed in Table 2, with the blood culture test used as the ‘gold standard’ for diagnosis of sepsis. Five infants had samples included from more than one episode of infection. To meet with laboratory regulations, samples that could be considered ‘high risk’ were excluded. Infants were not included in the study if the mother was known to be positive for hepatitis B, HIV or hepatitis C viruses. In cases where the mother was known to have a history of drug misuse and had not had antenatal screening for blood-borne viruses, the infants were also excluded. Other exclusion criteria were infants who did not require clinical blood samples and infants for whom extra blood sampling might be of particular risk, for example, infants with an underlying disorder causing anemia. Before embarking on this study we had previously performed a power calculation using the CodeLink chip platform [1] on neonatal samples but we also performed a power calculation using the Illumina chip platform, on an independent set of 30 infant samples at 9 months of age, before vaccination. This showed that the study design has 90% power to detect a twofold change in expression with an a of 1% (false discovery rate (FDR) corrected), for more than 99% of 35,177 gene probes present on the array [2]. A schematic of patient recruitment and sample processing workflow for the samples processed for the training, replication and validation arm of the study is shown in Fig. 1.
Table 1.
Patient demographics of samples used, microorganisms identified from infected patients and reasons for blood sampling in controls.
| Patient demographics of samples used | |||||||
|---|---|---|---|---|---|---|---|
| Sample set |
Training set |
Platform test set |
Validation test set |
||||
| Infection status | Infected (n = 28) | Control (n = 35) | Infected (n = 18) | Control (n = 24) | Infected (n = 16) | Control (n = 10) | |
| Male | 15 (54%) | 22 (63%) | 10 (56%) | 15 (63%) | 10 (63%) | 9 (90%) | |
| Gestation completed at birth (week): range (mean) | 24–38 (28.5) | 26–42 (37.9) | 24–38 (28.8) | 26–42 (37.3) | 23–40 (28.3) | 24–41 (31) | |
| Gestation completed at sampling (week): range (mean) | 26–39 (31.1) | 31–44 (39.4) | 26–39 (30.8) | 31–44 (39.1) | 25–41 (33.8) | 29–42 (34.9) | |
| Birthweight (g): range (mean) | 430–3380 (1126) | 650–4570 (3080) | 430–3380 (1236) | 650–4350 (2941) | 635–3160 (1134) | 800–4220 (1932) | |
| Microorganisms identified from infected patients |
Reasons for blood sampling in controls |
||||||
|---|---|---|---|---|---|---|---|
| Organism | Training set | Platform test set | Validation test set | Reason | Training set | Platform test set | Validation test set |
| Coagulase negative staphylococcus | 15 | 8 | 7 | Screening test: maternal thyroid disease | 17 | 9 | – |
| Enterococcus species | 4 | 3 | 1 | Bilirubin check due to jaundice | 5 | 4 | 1 |
| Group B Streptococcus | 2 | 2 | 1 | “Routine” neonatal screening (preterms) | 5 | 4 | 6 |
| Klebsiella species | 2 | 1 | 2 | Electrolyte check: previous deranged Na | 3 | 3 | – |
| Candida albicans and Klebsiella species | 1 | 1 | – | Screening test: pigmented scrotum | 2 | 1 | 3 |
| Escherichia Coli | 1 | 1 | 1 | Blood count check: Coomb's positive | 1 | 1 | – |
| Enterobacter cloacae | 1 | 1 | – | Screening test: newborn bloodspot | 1 | 1 | – |
| Pseudomonas aeruginosa | 1 | 1 | 1 | Neonatal encephalopathy | 1 | 1 | – |
| CMV | 1 | – | – | ||||
| Listeria monocytogenes | – | – | 1 | ||||
| Serratia marcescens | – | – | 2 | ||||
A. Patient demographics of samples used. Patient sample details are shown displaying the demographics of the population studied. B. Microorganisms identified from infected patients. Organisms detected for each infected infant are shown — these samples were taken at, or within 6 h of, the time of clinical suspicion of infection. C. Reasons for blood sampling in controls. The reasons for clinical blood sampling in the control group are shown — all of the screening tests in these infants were normal. Table 1 was adapted from Supplementary Table 3 of Smith et al. 2014 [2] by permission from Macmillan Publishers Ltd: Nature Communications [2], copyright (2014).
Table 2.
Clinical details of patient samples used in the study. Table 2 was adapted from Supplementary Data 4 of Smith et al. 2014 [2] by permission from Macmillan Publishers Ltd: Nature Communications [2], copyright (2014).
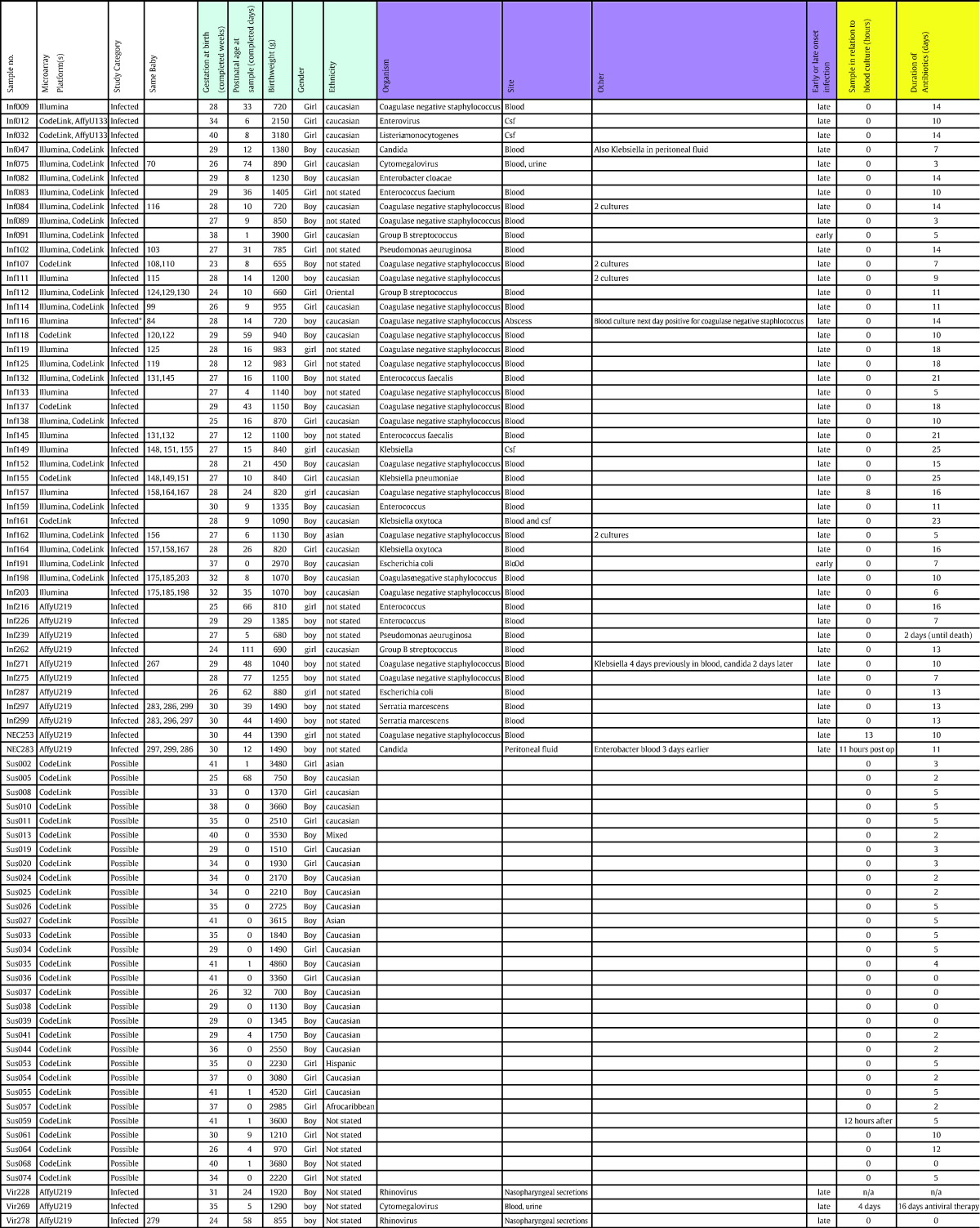

Fig. 1.
Study recruitment and sample processing. This flow diagram depicts process of neonatal subject recruitment over sample processing and microarray hybridization. Boxes and arrows are color-coded as follows. Healthy (presenting for clinical reasons other than suspected infection) control neonate samples = blue; neonate samples of suspected but unconfirmed infections = gray; neonate samples with blood-culture test confirmed infection = pink; neonate samples with blood-culture negative test but confirmed viral infection = striped pink. Figure 1 was adapted from Supplementary Figure 9 of Smith et al. 2014 [2] by permission from Macmillan Publishers Ltd: Nature Communications [2], copyright (2014).
Sample collection and RNA extraction
For RNA isolation, blood (500 μl–1 ml) was immediately injected into PAXgeneTM blood RNA tubes (PreAnalytiX BD/QIAgen) and mixed by inversion. Samples were then frozen at − 20 °C until RNA extraction occurred as described previously [1]. RNA was quantified and A260:A280 ratios generated using a ThermoSpectronic NanoDropTM1000 spectrophotometer. RNA quality was assessed qualitatively by examining the electropherogram and quantitatively from the RNA integrity number (RIN) generated by an Agilent 2100 Bioanalyser.
Gene expression profiling using microarrays
Microarray study design
This study was designed as a prospective case–control study to biologically and computationally infer a set of genes acting as a reliable classifier for bacterial sepsis in neonates. This design entails a main data set to identify genes and train a classification algorithm and is referred to as ‘training set’. Subsequent validation of the trained classifier requires independent data sets referred to as ‘test sets’ (distinctions between them outlined below). Based on earlier power calculations, the training set (Illumina HT-12 v3 platform) was established with 27 patient samples with a confirmed blood culture-positive test for sepsis (bacterial infected cases) and 35 age-matched controls (it also contains one cytomegalovirus-infected case that was not used for classification), all sub-selected by sample quality from the full study population. For assessing reproducibility of our gene classifier with a different assay platform, we examined a subset of 42 of these samples using the CodeLink gene expression platform (comprising 18 bacterial infected and 24 control samples) named in this study as ‘platform test set’. Subsequently, for independent clinical evaluation, the 52-gene set classifier was applied to a further 29 new and independent samples (comprising 16 bacterial infected, 3 viral infected and 10 control samples) named in this study as ‘validation test set’ which were analyzed using the CodeLink, Affymetrix HG-U133 Plus 2.0 and Affymetrix U219 gene expression platforms. The classifier was then used on 30 suspected infected samples and classification of samples into infected and non-infected cases compared against an ‘expert’ clinical classification.
RNA labeling and hybridization
For Illumina HT-12 v3 arrays total RNA was converted to double-stranded cDNA, followed by an in vitro transcription amplification step to generate labeled cRNA, using the Ambion Illumina TotalPrep-96 RNA Amplification Kit. The cRNA was quantified by A260 measurement using a NanoDropTM1000 spectrophotometer. The cRNA was normalized and hybridized onto the Illumina HT-12 v3 arrays overnight (16 h) at 58 °C. The unhybridized and non-specifically hybridized cRNA was washed away. The arrays are stained with Cy3-Streptavidin to bind to the analytical probes that have hybridized to the array. Arrays were scanned using an Illumina IScan scanner and fluorescence emissions were recorded in high-resolution images. The intensities of the images were extracted using GenomeStudio (2010.3) Gene Expression Module(1.8.0) software.
For CodeLink arrays the biotin-labeled cRNA target is prepared by a linear amplification method using tailed oligo dT priming of total RNA. After second-strand cDNA synthesis, the cDNA undergoes an in vitro transcription (IVT) reaction to produce the target cRNA. Various quality control procedures are incorporated. Hybridization is performed overnight and post-hybridization processing includes a stringent wash to remove unbound and non-specifically hybridized target molecules and staining with CyTM5-streptavidin conjugate. Several non-stringent washes remove unbound conjugate. The bioarrays are then dried and scanned on the Agilent G2567A scanner at 5 nm resolution. Raw data were obtained from the scanned images using CodeLinkTM EXPv4.1 (GE Healthcare) feature extraction software.
For Affymetrix HG-U133 plus 2.0 arrays biotin labeled cRNA target is prepared by a linear amplification method following reverse transcription of total RNA into T7 tailed double stranded cDNA. Biotinylated target cRNA was purified using RNeasy columns according to the manufacturer's instructions (QIAgen Ltd., Crawley, UK) and quantified by spectrophotometry. Fifteen micrograms of purified biotinylated cRNA was fragmented by heating for 35 min at 94 °C in the presence of magnesium ions, spiked with eukaryotic hybridization control and hybridized to HG-U133 plus 2.0 microarrays overnight at 45 °C. After hybridization the arrays were washed, stained with phycoerythrin coupled streptavidin and processed on the Affymetrix GeneChip Fluidics Workstation 400 using the EukGE-Ws2v4 protocol. Microarrays were then scanned using the Affymetrix GeneChip Scanner 3000 using GeneChip Operating Software instrument control and data acquisition system.
For Affymetrix U219 arrays total RNA was reverse transcribed to synthesize first-strand cDNA. This cDNA was then converted into a double-stranded DNA template for in vitro transcription to synthesize cRNA incorporating a biotin-conjugated nucleotide. This cRNA was then purified to remove unincorporated NTPs, salts, enzymes, and inorganic phosphate. The biotin-labeled cRNA was then fragmented and prepared for hybridization using the GeneChip HT Hybridization, Wash and Stain Kit for GeneTian (Affymetrix). Arrays were then processed and scanned on the Affymetrix GeneTitan Instrument as detailed in the Affymetrix GeneChip Command Console 2.0 User Guide.
Data normalization and analysis
For the computational and statistical pathway biology aspects of this study, a summary of the data analysis workflow is shown in Fig. 2. The chronological processing stages cover: data quality control, processing, statistical analysis, gene feature selection and classifier testing and validation.
Fig. 2.
Sequence of study analyses prior to validating 52-gene set as a classifier. This flow diagram identifies the sequence of analyses carried out on Illumina microarray data. The gray box indicates that the analyses within are used in combination to inform a subsequent result. Figure 2 was adapted from Supplementary Figure 10 of Smith et al. 2014 [2] by permission from Macmillan Publishers Ltd: Nature Communications [2], copyright (2014).
Data quality control: High-quality RNA (RNA integrity number (RIN) greater than 7) from infected and control infants were hybridized onto Illumina Human Whole-Genome Expression BeadChip HT-12 v3 microarrays comprising 48,802 features (human gene probes). Gene expression levels, distributions and controls were assessed using the arrayQualityMetrics package in Bioconductor [3]. A gender check was performed using Y-chromosome-specific loci.
Processing: Using the ‘lumi’ Bioconductor package, raw data from 63 samples were transformed using a variance stabilizing transformation before robust spline normalization to remove systematic between-sample variation. Microarray features that were not detected (using function ‘detectionCall’) on any of the arrays were removed from analysis and the remaining 23,342 features were used for subsequent statistical analysis.
Statistical analysis: Data were statistically examined to assess gestational age as a confounding factor. Within each sample group (control, infected), samples were age classified into bins based on the 33% and 66% corrected gestational age quantile values, yielding three age groupings. Per-gene hypotheses of differential expression between bacterial infection cases and control neonates were tested through linear modeling of the log2 scale expression values between groups and subsequent empirical Bayesian approaches to moderate the test statistic by pooling variance information from multiple genes (Bioconductor package ‘limma’ [4]). This included vertical p-value adjustment for multiple testing (Benjamini–Hochberg) to control for false discovery rate at a 1% level.
Gene feature selection for classifier: Computational network-based approaches were used to examine relationships in the data using correlation of gene expression and biological relationships. Statistically significant differentially expressed genes were examined further: heat maps and line graphs with hierarchical clustering by Euclidean distance were examined using Partek Genomics Suite v6.5, and visualization of networks of genes looking for patient-specific responses using BioLayout Express 3D [5]. These analyses were carried out step-wise using a pathway-biology approach, becoming more focused until a defined sub-network of 52 differentially expressed genes was identified [2]. The selected genes had adjusted p values of ≤ 10− 5, fold changes of ≥ 4 and were highly connected in terms of biological pathways and networks.
Classifier training and testing: First, a simulation model based on these 52 genes was established to assess the relationship between the number of gene predictors and classification error and establish suitability of this gene set for use with a panel of classifier algorithms. This approach used leave-one-out cross-validation with four different machine learning methods: Random Forests, Support Vector Machines, K Nearest Neighbour, and ROC-based [6], [7], [8], [9] (Fig. 3). Leave-one-out cross validation was repeated 100 times for each set of selected genes following a random ordering of the data at each replication to minimize variability of the error estimates [10].
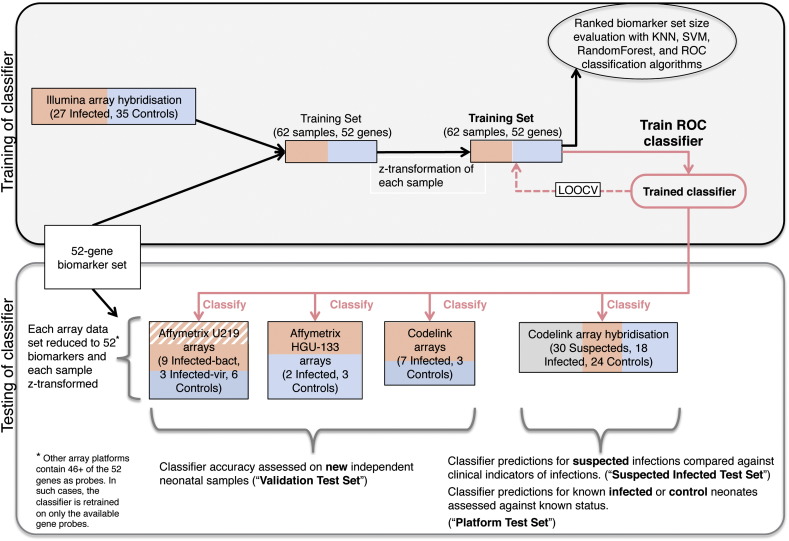
Fig. 3.
Training and testing of 52-gene classifier of sepsis in neonates. This diagram details the stages comprising training and testing of the ROC-based classifier. Top box represents processes in the training of the classifier; bottom box represents processes in the testing of the classifier on various types of test sets. LOOCV stands for leave-one-out-cross-validation, which is the iterative process in which a single sample of the training set is predicted based on the classifier trained on all remaining samples. Black arrows are data processing steps; red arrows indicate classifier training and prediction steps. Sample color coding: healthy (presenting for other clinical reasons than suspected infection) control neonate samples = blue; neonate samples of suspected but unconfirmed infections = gray; neonate samples with blood-culture test confirmed infection = pink; neonate samples with blood-culture negative test but confirmed viral infection = striped pink. Figure 3 was adapted from Supplementary Figure 11 of Smith et al. 2014 [2] by permission from Macmillan Publishers Ltd: Nature Communications [2], copyright (2014).
Next, the ROC-based classification method [9], (that does not require tuning of parameters and simplifies classification to a univariate decision that can easily be applied to independent data) was trained on the training set to learn the gene expression level differences that distinguish between controls and cases of infection. Internal accuracy of this classifier was tested through leave-one-out cross-validation on the training set, prior to its testing on independent data. Using the platform test set, a subset of 42 of the training set samples (18 infected, 24 controls) hybridized to CodeLink™ Whole Human Genome arrays, the trained ROC classifier was tested for platform-dependent performance. Subsequently, the classifier was tested for performance on completely new and independent neonatal samples, consisting of a further 26 samples (16 bacterially infected samples from 15 infants and ten control samples) which were run on CodeLink™ (seven infected, three control), Affymetrix® HG-U133 Plus 2.0 (two infected, three control) or Affymetrix® Human Genome U219 (nine infected, six control) arrays. Finally, the classifier was tested on n = 30 (hybridized to CodeLink arrays) new and independent cases where infection was suspected but not confirmed through blood culture and performance was compared against ‘expert’ clinical assessment (Table 2).
Discussion
We describe in this paper our detailed technical and analysis methodology for our data set describing the host response to neonatal infection. This data set is a unique repository of data describing the host response at the first sign of neonatal infection and has allowed identification of a 52-gene classifier that predicts bacterial infection with high accuracy. This data set lays the foundation for advancing diagnostic, prognostic and therapeutic strategies for neonatal sepsis and we hope will be of great value for future further investigations by the wider research community.
Conflict of interest
The authors declare there are no conflicting interests.
Acknowledgments
The authors would like to thank the infants and their parents for their participation in the study. This work was supported by the Wellcome Trust (WT066784) program grant, EU FP7 IAPP project ClouDx-i, Chief Scientists Office (ETM202) and BBSRC (BB/D019621/1) the Centre for Synthetic and Systems Biology at Edinburgh (SynthSys) supported by the BBSRC and EPSRC (BB/D019621/1) to P.G. and P.D.; MRC (G0701291) to K.L.F., P.D. and P.G. Teagasc (RMIS6018) funded D.J.L.'s participation in this study.
Contributor Information
Paul Dickinson, Email: paul.dickinson@ed.ac.uk.
Peter Ghazal, Email: p.ghazal@ed.ac.uk.
References
- 1.Smith C.L., Dickinson P., Forster T., Khondoker M., Craigon M., Ross A., Storm P., Burgess S., Lacaze P., Stenson B.J. Quantitative assessment of human whole blood RNA as a potential biomarker for infectious disease. Analyst. 2007;132:1200–1209. doi: 10.1039/b707122c. [DOI] [PubMed] [Google Scholar]
- 2.Smith C.L., Dickinson P., Forster T., Craigon M., Ross A., Khondoker M.R., France R., Ivens A., Lynn D.J., Orme J. Identification of a human neonatal immune-metabolic network associated with bacterial infection. Nat. Commun. 2014;5:4649. doi: 10.1038/ncomms5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kauffmann A., Gentleman R., Huber W. arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smyth G.K. Statistical Applications in Genetics and Molecular Biology Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microar. Stat. Appl. Genet. Mol. Biol. 2004;3(1):1–25. doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 5.Theocharidis A., van Dongen S., Enright A.J., Freeman T.C. Network visualization and analysis of gene expression data using BioLayout Express(3D) Nat. Protoc. 2009;4:1535–1550. doi: 10.1038/nprot.2009.177. [DOI] [PubMed] [Google Scholar]
- 6.Breiman L. Random Forests. Mach. Learn. 2001;45(1):5–32. [Google Scholar]
- 7.Cortes C., Vapnik V. Support-Vector Networks. Mach. Learn. 1995;20(3):273–297. [Google Scholar]
- 8.Altman N.S. An Introduction to Kernel and Nearest-Neighbour Nonparametric Regression. Am. Stat. 1992;46(3):175–185. [Google Scholar]
- 9.Lauss M., Frigyesi A., Ryden T., Hoglund M. Robust assignment of cancer subtypes from expression data using a uni-variate gene expression average as classifier. BMC Cancer. 2010;10:532. doi: 10.1186/1471-2407-10-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khondoker M.R., Bachmann T.T., Mewissen M., Dickinson P., Dobrzelecki B., Campbell C.J., Mount A.R., Walton A.J., Crain J., Schulze H. Multi-Factorial Analysis of Class Prediction Error: Estimating Optimal Number of Biomarkers for Various Classification Rules. J. Bioinf. Comp. Biol. 2010;8(6):945–965. doi: 10.1142/s0219720010005063. [DOI] [PubMed] [Google Scholar]