Abstract
The E2 protein is expressed in the early stage of human papillomavirus (HPV) infection that is associated with cervical lesions. This protein plays important roles in regulation of viral replication and transcription. To characterize the role of E2 protein in modulation of cellular gene expression in HPV infected cells, genome-wide expression profiling of human primary keratinocytes (HPK) harboring HPV16 E2 and HPV18 E2 was investigated using microarray. The Principle Components Analysis (PCA) revealed that the expression data of HPV16 E2 and HPV18 E2-transduced HPKs were rather closely clustered. The Venn diagram of modulated genes showed an overlap of 10 common genes in HPV16 E2 expressing HPK and HPV18 E2 expressing HPK. These genes were expressed with significant difference by comparison with control cells. In addition, the distinct sets of modulated genes were detected 14 and 34 genes in HPV16 E2 and HPV18 E2 expressing HPKs, respectively.
Keywords: HPV16 E2, HPV18 E2, Human primary keratinocytes
| Specifications | |
|---|---|
| Organism/cell line/tissue | Human primary keratinocyte (neonatal foreskin keratinocyte) |
| Sex | Male |
| Sequencer or array type | Microarray-Illumina Human HT-12 V4.0 expression BeadChip (Illumina, CA, USA) |
| Data format | Non-normalized data, normalized data and ANOVA in .text files |
| Experimental factors | GFP-HPV16/18 E2 expressing HPK vs. GFP expressing HPK |
| Experimental features | Keratinocytes were seeded and transduced with recombinant adenoviruses containing GFP, GFP-HPV16E2 or GFP-HPV18E2 at m.o.i. 50. The transduced cells were collected by scraping for RNA extraction after transduction 48 h. |
| Consent | n/a |
| Sample source location | n/a |
Direct link to deposited data
Deposited data are available here http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE54008.
Experimental design, materials and methods
Recombinant adenoviruses containing HPV E2s and transduction
Recombinant adenoviruses containing GFP, GFP-HPV16 E2 and GFP-HPV18 E2 were kindly provided by Thierry [1], [2] and used in this study. HPKs were seeded one day before infection. Adenovirus transduction was then conducted using recombinant adenoviruses containing GFP, GFP-HPV16E2 or GFP-HPV18E2 at multiplicity of infection (m.o.i.) of 50 [3] in quadruplicate experiments. The transduced cells were collected by scraping for RNA extraction after 48 h of transduction.
Total RNA isolation and microarray hybridization
Total RNA from recombinant adenoviruses transduced HPKs was isolated using RNeasy mini kit (QIAGEN, Singapore) followed by RNeasy mini method instructions. The total RNA was analyzed by RNA 6000 Nano Lab-on a-Chip in the 2100 Bioanalyzer (Agilent Technology, ON, CA), showing RIN score above 9.4. The total RNA was labeled and hybridized into one Human HT-12 v4.0 BeadChip (Illumina, CA, USA). The data files were quantified in GenomeStudio Version 2011.1 (Illumina, CA, USA) and exported for Partek Genomics Suite analysis.
Analysis of differential gene transcription
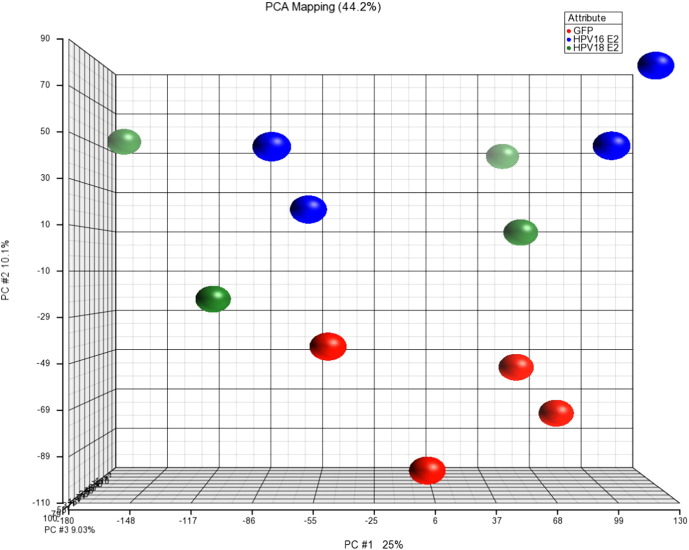
Gene expression data were imported into Partek Genomics Suite 6.5 (Partek, St Louis, MO, USA). Raw data were preprocessed, in steps of background correction, normalization using quantile normalization and summarization. The dataset were deposited at GSE54008 including GFP control (GSM1305807-10), GFP-HPV16 E2 (GSM1305811-14) and GFP-HPV18 E2 (GSM1305815-18). The Principal Components Analysis (PCA) was used for global visualization of all datasets (Fig. 1) [4]. The PCA analysis revealed the clear difference between GFP group and HPV E2 group. However, the HPV16 and HPV18 E2 groups showed overlapping correlation. Differential expression analysis of the samples was performed using one way analysis of variance (ANOVA). Gene lists were created using a cut-off of relative ± 1.5-fold change and p ≤ 0.05. The Venn diagram of modulated genes in HPV16 and 18 E2 expressing HPK was created. The result showed that 10 genes were commonly found in both HPV16 and HPV18 E2 expressing HPKs whereas the mutually exclusive genes in each HPV16 E2 and HPV18 E2 expressing HPKs were 14 and 34 genes, respectively (Fig. 2). The HPV16 E2 dataset was previously used for immune associated gene analysis in a published work [5].
Fig. 1.
The Principal Components Analysis of dataset. The GFP, GFP-HPV16 E2 and GFP-HPV18 E2 dataset were analyzed to identify the outlier of sample group. Red; GFP group, blue; GFP-HPV16 E2 group, and green; HPV18 E2 group. Each group was represented in 4 replicates.
Fig. 2.
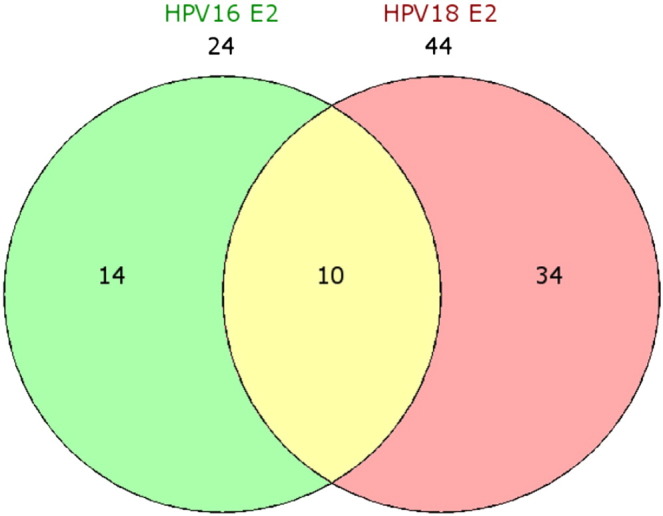
The Venn diagram of modulated genes in HPV16 and 18 E2 expressing HPK. The result showed 10 genes commonly modulated by both HPV16 and HPV18 E2 expressing HPKs (yellow area) and mutually exclusive modulated genes in each HPV16 E2 and HPV18 E2 expressing HPK were 14 (green area) and 34 genes (pink area), respectively.
Conclusion
This study generated the dataset from the microarray analysis of high-risk HPV16 and HPV18 E2-expressing HPKs which might be useful to gain further insights into the mechanisms underlying the crosstalk between high-risk HPV E2 and host cells.
Conflict of interest
The authors have no conflicts of interest.
References
- 1.Demeret C., Garcia-Carranca A., Thierry F. Transcription-independent triggering of the extrinsic pathway of apoptosis by human papillomavirus 18 E2 protein. Oncogene. 2003;22:168–175. doi: 10.1038/sj.onc.1206108. [DOI] [PubMed] [Google Scholar]
- 2.Blachon S., Bellanger S., Demeret C., Thierry F. Nucleo-cytoplasmic shuttling of high risk human papillomavirus E2 proteins induces apoptosis. J. Biol. Chem. 2005;280:36088–36098. doi: 10.1074/jbc.M505138200. [DOI] [PubMed] [Google Scholar]
- 3.Xue Y., Lim D., Zhi L., He P., Abastado J.P., Thierry F. Loss of HPV16 E2 protein expression without disruption of the E2 ORF correlates with carcinogenic progression. Open Virol. J. 2012;6:163–172. doi: 10.2174/1874357901206010163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raychaudhuri S., Stuart J.M., Altman R.B. Principal components analysis to summarize microarray experiments: application to sporulation time series. Pac. Symp. Biocomput. 2000:455–466. doi: 10.1142/9789814447331_0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sunthamala N., Thierry F., Teissier S., Pientong C., Kongyingyoes B., Tangsiriwatthana T., Sangkomkamhang U., Ekalaksananan T. E2 proteins of high risk human papillomaviruses down-modulate STING and IFN-kappa transcription in keratinocytes. PLoS One. 2014;9:e91473. doi: 10.1371/journal.pone.0091473. [DOI] [PMC free article] [PubMed] [Google Scholar]