Abstract
Given the recognized role of the commensal microbiota in regulating host immunity to pathogens, it is not surprising that microbiota are also capable of regulating autoimmune responses. The underlying mechanisms of autoimmune regulation by the microbiota are just beginning to emerge. Here, we discuss possible pressure points towards the development of autoimmune diseases that can be influenced by the microbiota. Besides acting on the adaptive and innate arms of the immune response, the microbiota can affect the targets of autoimmunity directly, even during development in utero, and be involved in regulation of autoimmunity via interactions with hormones.
Introduction
Research of the past decade has helped us appreciate the importance of the commensal microbiota in autoimmune disease development (Chervonsky, 2010). Along the way, the difficulties in understanding the mechanisms by which microbes influence autoimmune diseases pathogenesis have also become apparent. The confounding factors to understanding the underlying mechanisms include: a) the enormous complexity of the microbiota; b) involvement of microbial communities rather than isolated lineages in interactions with the host; c) sensitivity of the microbiota to various environmental insults including dietary, chemical and biological; d) the multiplicity of host microbial sensors; and e) the difficulties in applying classical Koch’s postulates to commensal microbes. The revival of the gnotobiotic approach (colonization of germ-free animals with defined microorganisms or their consortia) and its application in the context of animal models of autoimmunity have helped alleviate some of the constraints noted above and have led to several important conclusions. Here, we review recent findings that link the microbiota to the development of autoimmune disease and discuss the points where microbiota can potentially apply pressure on the path to autoimmune development, drawing specific examples from the literature where available.
Exploring microbial connections to autoimmunity
Mouse models of human monogenic diseases such as immunodysregulation polyendocrinopathy enteropathy X-linked syndrome (IPEX) and autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APADEC) -- mice harboring Foxp3 and AIRE lesions, respectively -- were shown to be independent of microbiota, whereas models of polygenic diseases such as Type 1 diabetes (T1D), rheumatoid arthritis (RA) or multiple sclerosis (MS) were found to be affected by the microbiota. Importantly, some autoimmune conditions such as T1D in predisposed non-obese diabetic (NOD) mice and BB rats developed without the presence of microbes, whereas others were either augmented by microbes or required their presence for disease onset. These results imply some level of specificity in the ability of microbial lineages to induce different diseases. For example, mono-colonization with segmented filamentous bacteria (SFB) augmented rheumatoid arthritis (RA) in a mouse model (Wu et al., 2010), but had no effect in the context of T1D (Yurkovetskiy et al, 2013). However, the variability in the genomes of bacteria commonly identified as SFB could explain why this bacterium was protective in an earlier T1D study (Kriegel et al, 2011). Despite these complications we have learned that microbes can be protective, neutral or provocative for the development of autoimmunity. The gnotobiotic approach can potentially be used to study the effects of the human and animal microbiota on animal pathology. However, it is not a high throughput method and the few available animal models are unlikely to fully represent the diverse human pathologies associated with Systemic Lupus Erythematosus (SLE) or T1D. On the other hand, high throughput methods of microbiota analysis, including sequencing of 16S rRNA genes, whole microbial genome sequencing, and metabolomic analysis, have enabled studies with patients and matching control human populations to assess associations between autoimmune disease and microbial communities. For example, studies on three human cohorts that included T1D patients have found links between microbiota composition and autoimmune disease. All three studies were done in Scandinavia where the incidence of T1D is high. However, the cohorts analyzed were relatively small [between 8 and 36 subjects including healthy controls (Brown et al., 2011, de Goffua et al., 2013, Kostic et al., 2015)]. For comparison, genome-wide association studies (GWAS) of T1D patients usually involve thousands of cases and samples allowing the identification of rather small contributions of individual genes to disease development. Not surprisingly, the microbial associations with T1D differed in these studies. Brown et al. and De Goffau et al. identified a decrease in lactate- and butyrate-producing bacteria as being associated with development of autoimmunity, whereas Kostic et al. did not see similar changes in patients that developed autoimmunity during the study period. Differences in methods (sequencing bias due to different read depths or different hypervariable regions of 16S rRNA amplification used for analysis) and study groups (patients with established T1D vs. patients with anti-pancreatic antibodies who later develop overt diabetes) may account for the differences in the results. Importantly, all studies repeatedly found that diabetic patients had one common trend – the alpha-diversity of their microbiota was reduced. The reduction of microbial diversity, usually referred to as dysbiosis, is also a feature of Inflammatory Bowel Disease (IBD) (see review in this issue by Wlodarska et al.), suggesting the existence of important underlying commonalities between very different inflammatory conditions.
The microbiota is extremely sensitive to dietary changes (Wu et al., 2011; Carmody et al., 2015) and likely also to changes in the host’s metabolism. Given that T1D significantly affects host metabolism, microbial community alterations observed in T1D patients could actually be dictated by the disease itself (Kostic et al., 2015). Indeed, recent studies have suggested that changes in stool metabolites (caused by changes in microbiota composition) can serve as biomarkers for predicting overt diabetes (Scher et al., 2013; Kostic et al., 2015). Finding a correlation between the composition of microbial communities and autoimmune diseases and eventual proof of their diagnostic or therapeutic value constitute an important part of the research in this area. This requires prospective and longitudinal studies with larger cohorts of patients, such as the TEDDY study (Hagopian et al., 2006).
The discovery of specific activating and inhibitory receptors triggered by the microbiota and engaged in autoimmunity is another avenue towards understanding the microbiota-autoimmunity connection. These studies can produce important information even without a complete knowledge of the microbial perpetrator’s identity.
Provision of ligands for adaptive immunity
It has long been known that pathogenic microbes can potentially provide ligands that elicit cross-reactivity with self antigens (molecular mimicry) (Oldstone, 1987; Wucherpfennig and Strominger, 1995), and this has also been proposed as a possible link between commensal microbiota and autoimmunity (Chervonsky, 2009). Novel approaches involving MHC-peptide tetramers in humans (Su et al., 2013) and mice (Nelson et al., 2014) have provided concrete evidence for the existence of T cell receptors (TCR) that cross-react to peptides that the host was not previously exposed to. Synthetic peptides derived from microbes were able to activate cells carrying such TCRs. Although these results provide a solid basis for further experiments that could prove that TCR promiscuity leads to autoimmunity, direct proof of this occurring in vivo remains outstanding. For homologous or even identical peptides to elicit a T cell response, they have to become available to MHC-expressing antigen presenting cells (APC) and be properly processed by proteases and by the peptide-loading machinery. It is not impossible that the reduction of microbial diversity associated with inflammatory and autoimmune diseases (Brown et al., 2011; Scher et al., 2013) creates conditions under which microbes or their products can more efficiently cross the epithelial barriers and be appreciated by APCs. Solid evidence for the role of commensals in providing antigens for the activation of the host’s cross-reactive T cells is desperately needed. In that regard, the recent finding that arthritis-promoting SFB (Wu et al., 2010) also elicit Th17 T cell responses against themselves (Yang et al., 2014) is very intriguing. It remains to be seen if molecular mimicry is involved in the modulation of autoimmunity by these bacteria.
Influencing the cells of the immune system
The host immune system plays an important role in shaping the microbiota and reciprocally, host-associated microbes significantly influence the development and function of innate and adaptive immunity (reviewed by Hooper et al., 2012). Commensal microbes are sensed by multiple innate immune sensors. Signaling though innate immune receptors ultimately leads to activation of adaptive immune responses. Thus, microbes are likely to challenge the activation of adaptive autoimmunity by interfering with innate-adaptive communication (Chervonsky, 2010). Cells of the innate immune system such as dendritic cells, macrophages, neutrophils and NK cells are necessary for the development of many types of autoimmunity and microbes can also influence the activity of these types of cells. Macrophages isolated from germ free mice release lower levels of TNF-α and higher levels of IL-10 when stimulated with bacterial lipopolysaccharide (LPS) compared to macrophages from specific pathogen free (SPF) mice (Souza et al., 2010). Though total numbers of splenic dendritic cells are not much different between germ free and SPF animals, microbial activation of these cells leads to decreased expression of IL-15, IL-6, TNF-α and type I interferons. NK function is also impaired under germ free conditions, possibly due to the inability of monocyte-derived cells to produce type I interferons in response to microbial stimuli (Ganal et al., 2012). The proper function and activity of bone marrow-derived neutrophils is highly influenced by exposure to commensal organisms, likely in the form of circulating microbial products such as muramyl dipeptide (MDP) (Clarke et al., 2010). iNKT cells, which also contribute to several autoimmune diseases, have some defects in the germ free environment and require microbial ligands for their maturation (Wingender et al., 2012; An et al., 2014). Germ free mice are reported to also have distinct alterations in their adaptive immune system including decreased numbers of T and B cells, decreased levels of IgA and IgG antibodies, and a strong skewing towards the Th2 CD4+ T cell helper subset. Introduction of microbes to germ free mice restores the Th1 and Th17 compartments. Development of inducible regulatory T cells (Tregs) in the periphery is dependent on interactions with the microbiota and is influenced by microbially produced short chain fatty acids (Arpaia et al., 2013). Tregs do have a measurable degree of control over developing autoimmunity because their ablation in T1D-susceptible mice leads to immediate overt diabetes (Feuerer et al., 2009). The programming of both innate (Ganal et al., 2012) and adaptive cells (Arpaia et al., 2013) by microbial products is likely mediated through epigenetic changes at transcription factor target sites.
Indirect modulation of autoimmunity
In addition to the well-appreciated direct effects of microbiota on the immune system as discussed above, there are also indirect ways for microbiota to affect autoimmunity.
a. Affecting the target organs
GWAS evidence suggests that organs targeted in autoimmunity may not just be passive bystanders in the face of immune-mediated attack (Santin and Eizirik 2013; Baranzini et al., 2013). Microbial metabolites, which are heavily dependent on consumed nutrients, could serve as links between the microbiota and the host’s organs. One can envision a role of microbial metabolites in regulating the stress experienced by the target organ, either easing it or exacerbating it. For example, the increased resistance of pancreatic beta cells to stress could reduce apoptosis and the supply of autoantigens, thus reducing the pace or magnitude of autoreactive beta cell-specific T cell activation. The resulting slow kinetics of activation may also provide a better chance for Tregs to exert immune control.
Diet is undoubtedly very important in influencing the metabolic functions of the microbiota and the host (Scott et al., 2013; Carmody et al., 2015). Since diet-induced shifts in the microbiota may have a role in development of autoimmunity, it is tempting to speculate that the beneficial impact of dietary intervention (for example in T1D) is due to changes in the microbiota. However, a recent study in T1D-predisposed BB-rats (Patrick et al., 2013) identified a semi-purified diet that was protective against T1D in both SPF and germ free conditions, indicating that the protection against disease was independent of the microbiota. Whether this type of dietary intervention has a direct impact on beta cell function is unknown. Interestingly, a recent clinical trial of a similarly designed baby formula failed to provide significant protection from anti-pancreatic antibody development (Knip et al., 2014). Clearly there are still many unknown factors, including the role of the microbiota, which could influence the outcome of such a treatment.
b. Developmental influences
Unlike invertebrates such as Drosophila or hydra (Shin et al., 2011; Rahat and Dimentman, 1982), mammalian development does not seem to be directly and irreversibly influenced by microbiota (with the important note that developing germ free animals are always artificially provided with necessary nutrients). That seems to also be true for the immune system. Although germ free animals have an underdeveloped mucosa-associated lymphoid tissue and less activated local and global adaptive immune system (Sommer and Backhed, 2013), their immune functions can be restored to apparently normal states by colonization with commensal microbes (Smith et al., 2007).
Nevertheless, the environment experienced both during gestation and during early life development may lead to phenotypic states different from what genetics alone would predict. Though there is a paucity of evidence for transgenerational imprinting on the function of the immune system, there is mounting evidence that malnutrition and parental experiences such as stress and obesity can contribute to metabolic disease development in the offspring (Aiken and Ozanne, 2014; Radford et al 2014). Since the microbiota can contribute to metabolic dysfunction, it may be considered an environmental factor in transgenerational extra-genetic phenotype programming. In the few experiments relevant to autoimmunity, exposure of NOD mice to a special diet formulation until weaning age was sufficient to decrease the incidence of T1D, as long as these animals were also exposed to the same diet in utero (Kagohashi et al., 2006).
Another dietary intervention during gestation, maternal exposure to gluten, affected the development of T1D: the progeny of NOD mothers fed gluten-free diet during pregnancy and exposed to gluten-containing chow throughout their life had a substantially decreased T1D incidence (Hansen et al., 2014). The potential role of the microbiota in this process has not been addressed. In a different experiment, maternal environment has also been shown to affect the development of T1D, as embryos transplanted from NOD mice to DBA females were protected from development of the disease after birth (Greeley et al., 2002). Thus, contributions from the maternal environment may play a role in shaping microbiota composition and thereby influence the risk for disease development.
Pregnancy itself imposes changes upon the intestinal microbiota in humans: the third trimester microbiota induced greater adiposity and insulin resistance when transferred to germ free animals compared to microbiota at the first trimester (Koren et al., 2012). The risk for development of T1D may thus be linked to the imprinting of metabolic functions on the insulin-producing beta cells.
Long-lasting imprinting effects may not necessarily be affecting the target organs alone, but also the developing immune system. In this regard, it is important to note that several populations of immune cells have been recently found to be long-living and embryonically-derived, such as tissue-resident macrophages and B1 B cells (Gomez Perdiguero et al., 2015, Montecino-Rodriguez and Dorshkind, 2012]. These cell types are likely to be affected by the microbiota and related metabolic cues during development and later contribute to the overall status of the immune system and responses to self and foreign antigens.
c. Hormones and microbes
Sexual dimorphism is an important aspect of many autoimmune diseases and an unexpected role for the microbiota in mediating this sexual dimorphism has been recently uncovered. Previous experiments have suggested that male hormones are protective in SLE or T1D and estrogens may contribute to disease progression (reviewed in Markle and Fish, 2014). Two recent studies have connected hormonal influences and microbiota to explain the sexual dimorphism of autoimmunity (Markle et al., 2013; Yurkovetskiy et al., 2013). They were based on previous findings that germ free animals lose the sexual dimorphism of T1D, with both females and males having a high incidence of the disease. Both studies found that microbiota between male and female littermates differ after puberty and that the microbiota contributed to increased levels of testosterone in the blood. The microbiota of castrated males was more similar to the microbiota of post-pubescent females than to that of post-pubescent males. However, overall these studies did not reveal a gender-specific microbiota signature, as the differences in bacterial composition between males and females did not overlap in multiple sequencing experiments. While the two studies concur that colonization with microbes increases the levels of androgens in the blood, they differed in their interpretation of the results. Markle et al. found that the transfer of male microbiota to females was protective and suggested a linear model wherein microbiota induced testosterone which then affects autoimmunity. Based on the lack of a correlation between the levels of microbially-induced testosterone and the ability of particular microbes to prevent T1D, Yurkovetskiy et al. suggested a dual signal hypothesis. In this hypothesis signals from both hormones and microbiota were required to ensure male protection from T1D. Interestingly, in gnotobiotic experiments, male protection (but not female protection) was achieved by mono-colonization with bacteria of entirely unrelated genera. Bacteria as different as SFB and an E.coli-like species both induced protection in males. This result also suggested the existence of multiple mechanisms of microbial involvement in promoting sexual dimorphism.
Whether microbes contribute to sexual dimorphism in other autoimmunity models remains to be examined. Investigation of the role of the microbiota in biasing autoimmunity in humans may be more productive if it is not solely based on microbiota surveys, but on the translation of discoveries of signaling pathways regulated by the endocrine system and microbes in animal models.
Concluding remarks
We have suggested several underappreciated contact points where the microbiota can exert its influence on inflammatory and autoimmune diseases. Additionally, several viruses were recently found to depend on the microbiota for their propagation. Remarkably, T1D induction in rats by Kilham virus was sensitive to antibiotics (Hara et al., 2012). The involvement of viruses and their linkage with bacterial commensals further complicate the understanding of the microbiota’s role in autoimmunity.
Needless to say, the overall picture has proven to be more complex than the original linear interpretations of the microbiota’s effects. To add to the complexity, many effects are likely to be species-specific and direct translation from rodents to humans and back must be made with caution. The well-established anti-inflammatory effects of Bacteroides fragilis have been observed in mice, but this commensal is known to cause inflammatory responses in humans. SFB is another example that has defined pro- and anti-autoimmune effects in mice, but does not have an obvious human counterpart. These difficulties should not discourage future investigations, because although microbes involved in autoimmunity could be different between species due to genetic control, diet and life style, the principles of microbial participation and pathway regulation are most likely very similar.
Contemporary methods of high throughput analysis of microbial communities and their metabolomes, in combination with classical bacterial and mammalian genetics should be able to identify the critical pathways and bring us closer to knowledge-based therapies.
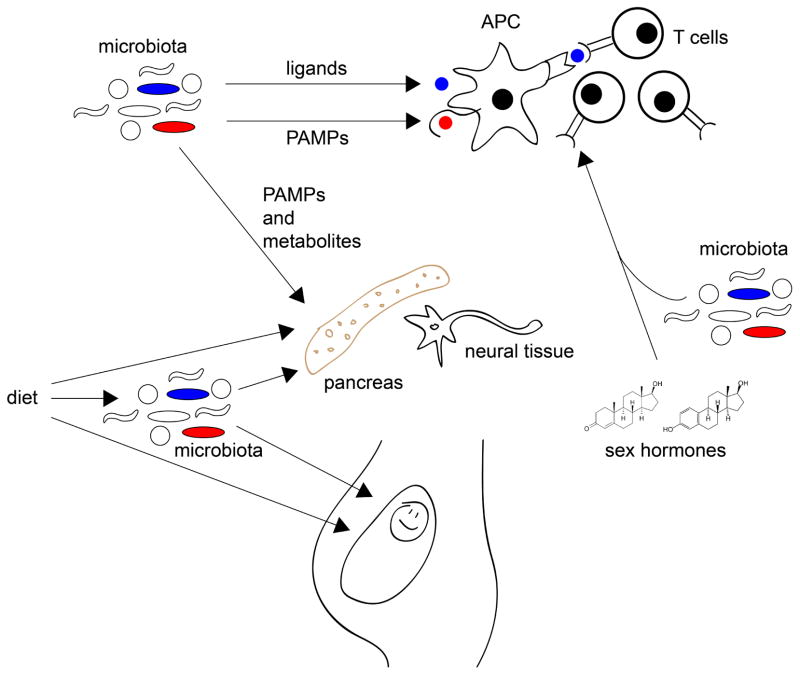
Figure. Environment, diet and microbes may influence the development of autoimmunity at various stages of life.
The microbiota may impinge on autoimmunity in many ways. It can be a source of peptide ligands for T cell recognition, structural components (Pathogen-Associated Molecular Patterns, PAMPs) recognized by innate immune receptors, and metabolites that can serve as an energy source or regulate cellular biosynthetic and signaling pathways. Besides direct effects on the adaptive and innate immune systems), microbial PAMPs and metabolites can affect the target organs (such as pancreas or neural tissue), changing the levels of stress experienced by these organs and thus affecting the pace with which autoimmunity is activated. Diet can affect target organs directly or through the microbiota. These influences do not start after birth, but may occur in the mother’s womb, predisposing the progeny to disease. The microbiota also cooperates with the endocrine system to drive sexual dimorphism, a prominent feature of the major autoimmune conditions.
Microbiota can regulate autoimmune responses. Yurkovetskiy et al. discuss possible pressure points towards the development of autoimmune diseases that can be influenced by the microbiota. Besides acting on the immune system, microbiota can affect the targets of autoimmunity directly and be involved in regulation of autoimmunity via interactions with hormones.
Acknowledgments
LAY was supported by NIH T32 GM007183. JMP was supported by NIH T32 AI007090. AVC is supported by NIH/NIDDK Digestive Disease Research Core Center grant DK42086, NIH grant AI082418, and by JDRF grant 17-2011-519.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aiken CE, Ozanne SE. Transgenerational developmental programming. Human reproduction update. 2014;20:63–75. doi: 10.1093/humupd/dmt043. [DOI] [PubMed] [Google Scholar]
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, Kasper DL. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, Casella G, Drew JC, Ilonen J, Knip M, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PloS one. 2011;6:e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, Gerber GK, Luevano JM, Jr, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. Diet dominates host genotype in shaping the murine gut microbiota. Cell host & microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervonsky A. Innate receptors and microbes in induction of autoimmunity. Current opinion in immunology. 2009;21:641–647. doi: 10.1016/j.coi.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervonsky AV. Influence of microbial environment on autoimmunity. Nature immunology. 2010;11:28–35. doi: 10.1038/ni.1801. [DOI] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nature medicine. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Goffau MC, Luopajarvi K, Knip M, Ilonen J, Ruohtula T, Harkonen T, Orivuori L, Hakala S, Welling GW, Harmsen HJ, et al. Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes. 2013;62:1238–1244. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Shen Y, Littman DR, Benoist C, Mathis D. How punctual ablation of regulatory T cells unleashes an autoimmune lesion within the pancreatic islets. Immunity. 2009;31:654–664. doi: 10.1016/j.immuni.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greeley SA, Katsumata M, Yu L, Eisenbarth GS, Moore DJ, Goodarzi H, Barker CF, Naji A, Noorchashm H. Elimination of maternally transmitted autoantibodies prevents diabetes in nonobese diabetic mice. Nature medicine. 2002;8:399–402. doi: 10.1038/nm0402-399. [DOI] [PubMed] [Google Scholar]
- Hagopian WA, Lernmark A, Rewers MJ, Simell OG, She JX, Ziegler AG, Krischer JP, Akolkar B. TEDDY--The Environmental Determinants of Diabetes in the Young: an observational clinical trial. Annals of the New York Academy of Sciences. 2006;1079:320–326. doi: 10.1196/annals.1375.049. [DOI] [PubMed] [Google Scholar]
- Hansen CH, Krych L, Buschard K, Metzdorff SB, Nellemann C, Hansen LH, Nielsen DS, Frokiaer H, Skov S, Hansen AK. A maternal gluten-free diet reduces inflammation and diabetes incidence in the offspring of NOD mice. Diabetes. 2014;63:2821–2832. doi: 10.2337/db13-1612. [DOI] [PubMed] [Google Scholar]
- Hara N, Alkanani AK, Ir D, Robertson CE, Wagner BD, Frank DN, Zipris D. Prevention of virus-induced type 1 diabetes with antibiotic therapy. Journal of immunology. 2012;189:3805–3814. doi: 10.4049/jimmunol.1201257. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium. Network-based multiple sclerosis pathway analysis with GWAS data from 15,000 cases and 30,000 controls. American journal of human genetics. 2013;92:854–865. doi: 10.1016/j.ajhg.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagohashi Y, Tatewaki R, Takebe E, Naora H, Abiru N, Kobayashi M, Hashimoto M, Shido O, Otani H. Maternal EFA composition affects the pathogenesis of type 1 diabetes in NOD mouse offspring. Zool Sci. 2006;23:1221–1221. [Google Scholar]
- Knip M, Akerblom HK, Becker D, Dosch HM, Dupre J, Fraser W, Howard N, Ilonen J, Krischer JP, Kordonouri O, et al. Hydrolyzed infant formula and early beta-cell autoimmunity: a randomized clinical trial. Jama. 2014;311:2279–2287. doi: 10.1001/jama.2014.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, Hamalainen AM, Peet A, Tillmann V, Poho P, Mattila I, et al. The Dynamics of the Human Infant Gut Microbiome in Development and in Progression toward Type 1 Diabetes. Cell host & microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle JG, Fish EN. SeXX matters in immunity. Trends in immunology. 2014;35:97–104. doi: 10.1016/j.it.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- Montecino-Rodriguez E, Dorshkind K. B-1 B cell development in the fetus and adult. Immunity. 2012;36:13–21. doi: 10.1016/j.immuni.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RW, Beisang D, Tubo NJ, Dileepan T, Wiesner DL, Nielsen K, Wuthrich M, Klein BS, Kotov DI, Spanier JA, et al. T Cell Receptor Cross-Reactivity between Similar Foreign and Self Peptides Influences Naive Cell Population Size and Autoimmunity. Immunity. 2014 doi: 10.1016/j.immuni.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone MB. Molecular mimicry and autoimmune disease. Cell. 1987;50:819–820. doi: 10.1016/0092-8674(87)90507-1. [DOI] [PubMed] [Google Scholar]
- Patrick C, Wang GS, Lefebvre DE, Crookshank JA, Sonier B, Eberhard C, Mojibian M, Kennedy CR, Brooks SP, Kalmokoff ML, et al. Promotion of autoimmune diabetes by cereal diet in the presence or absence of microbes associated with gut immune activation, regulatory imbalance, and altered cathelicidin antimicrobial Peptide. Diabetes. 2013;62:2036–2047. doi: 10.2337/db12-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford EJ, Ito M, Shi H, Corish JA, Yamazawa K, Isganaitis E, Seisenberger S, Hore TA, Reik W, Erkek S, et al. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science. 2014;345:1255903. doi: 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahat M, Dimentman C. Cultivation of bacteria-free Hydra viridis: missing budding factor in nonsymbiotic hydra. Science. 1982;216:67–68. doi: 10.1126/science.7063873. [DOI] [PubMed] [Google Scholar]
- Santin I, Eizirik DL. Candidate genes for type 1 diabetes modulate pancreatic islet inflammation and beta-cell apoptosis. Diabetes, obesity & metabolism. 2013;15(Suppl 3):71–81. doi: 10.1111/dom.12162. [DOI] [PubMed] [Google Scholar]
- Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol Res. 2013;69:52–60. doi: 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Seminars in immunology. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Sommer F, Backhed F. The gut microbiota--masters of host development and physiology. Nature reviews Microbiology. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Souza DG, Vieira AT, Soares AC, Pinho V, Nicoli JR, Vieira LQ, Teixeira MM. The essential role of the intestinal microbiota in facilitating acute inflammatory responses. Journal of immunology. 2004;173:4137–4146. doi: 10.4049/jimmunol.173.6.4137. [DOI] [PubMed] [Google Scholar]
- Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38:373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Kostic AD, Xavier RJ. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe. 2015 doi: 10.1016/j.chom.2015.04.008. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender G, Stepniak D, Krebs P, Lin L, McBride S, Wei B, Braun J, Mazmanian SK, Kronenberg M. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012;143:418–428. doi: 10.1053/j.gastro.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucherpfennig KW, Strominger JL. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]