Abstract
To comprehensively reflect the roles of Rpl11 on the transcriptome of zebrafish model of Diamond-Blackfan Anemia (DBA), we performed whole-genome transcriptome sequencing on the Illumina Hi-Seq 2000 sequencing platform. Two different transcriptomes of zebrafish Rpl11-deficient and control Morpholino (Mo) embryos were collected and analyzed. The experimental design and methods, including sample preparation, RNA-Seq data evaluation and treatment, were described in details so that representative high-throughput sequencing data were acquired for assessing the actual impacts of Rpl11 on zebrafish embryos. We provided the accession number GSE51326 for easy access to the database.
Keywords: Transcriptome, Zebrafish, DBA, High-Seq 2000, IPA
| Specifications | |
|---|---|
| Organism/cell line/tissue | Zebrafish embryos |
| Sex | N/A |
| Sequencer or array type | Illumina High-Seq 2000 |
| Data format | Raw data |
| Experimental factors | Zebrafish embryos at the one-cell stage were injected with Rpl11 Mo or control Mo using a microinjector |
| Experimental features | Sample preparation of zebrafish embryos at 48 hpf, whole-genome transcriptome sequencing, RNA-Seq data evaluation and treatment. |
| Consent | N/A |
| Sample source location | All zebrafish embryos were collected in Huazhong University of Science and Technology. |
Direct link to deposited data
Deposited data can be found here: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=mzcbuyyipbixhun&acc=GSE51326.
Experimental design, materials and methods
Sample preparation
To obtain the representative whole-genome transcriptome sequencing data, sample preparation is one of the critical steps in this study. Firstly, as to the dosage of Mo injected in the zebrafish embryos, we compared different dosages of Rpl11 Mo, together with control Mo, which has a few nucleotide mutations, to reduce the non-specific effects of rpl11 Mo. 0.5 ng Mo/each embryo was found to be sufficient to completely inhibit GFP-Rpl11 fusion gene expression. This dosage is too low to produce off target effect. Secondly, as to the generated Rpl11-knockdown zebrafish embryos, we observed the obvious hematopoietic defects of Rpl11-deficient zebrafish embryos as early as the start of hematopoietic 48 hpf by comparing with that of Mo ctrl. At this stage, the zebrafish primitive hematopoiesis and definitive hematopoiesis wave appeared. We collected zebrafish embryos with obviously hematopoietic defects for high-throughput sequencing. Thirdly, to increase the reproducibility of the experimental results, we selected about 40–50 zebrafish embryos from at least three experiments and mixed them together to obtain the representative biological repeat. This strategy was also used in previous high-throughput sequencing studies [1], [2].
Transcriptome sequencing
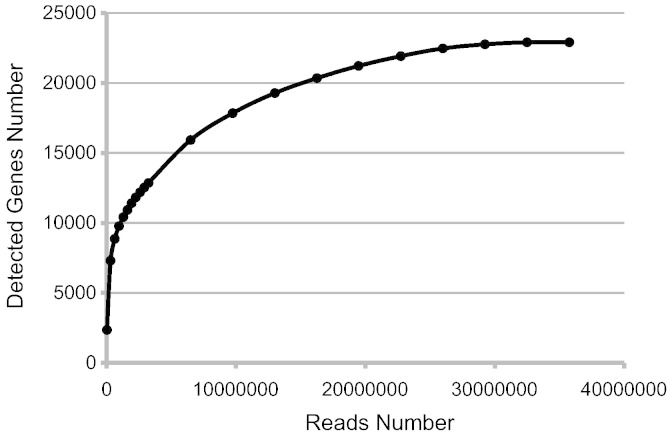
To obtain the representative data, the manipulations for two samples of Rpl11 Mo and control Mo, including RNA extraction, mRNA-library construction (Illumina mRNA-Seq library preparation kit) and sequencing on the High-Seq 2000 platform, were carried out in parallel completely. Paired-end sequencing mode (2 × 100 bp) was employed to match more accurately the reference genome sequence and improve sequencing efficiency. Moreover, pair-end sequencing data has more biologically analytical options including alternative splicing analysis [3]. In this study, 4–6 G of sequencing data was finally collected. The present sequencing data was completely saturated [4] and sufficient for subsequent analysis (Fig. 1).
Fig 1.
Saturation curve of transcriptomic sequencing reads for zebrafish samples.
Hi-Seq data processing
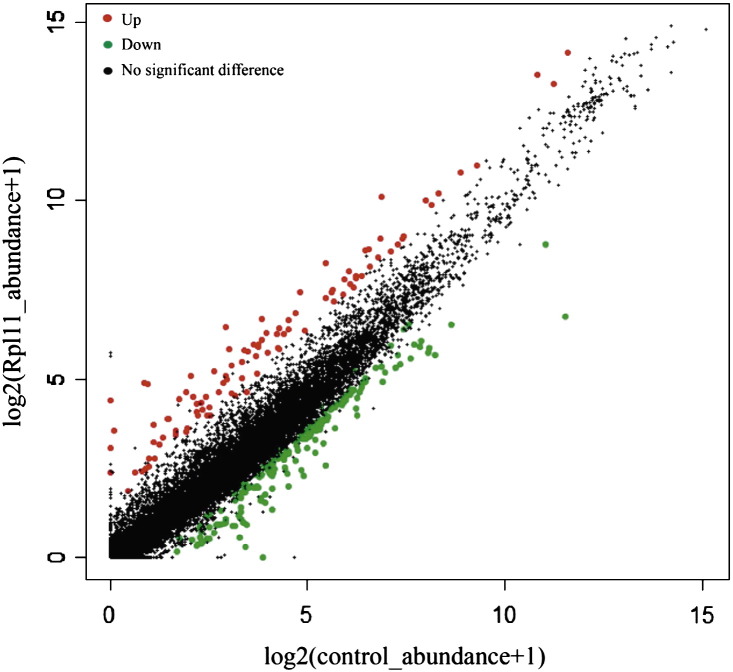
The collected transcriptome sequencing data was mapped to the latest version of the zebrafish genome (Zv9). More than 10,000 gene transcripts were mapped to known zebrafish genes, which is similar to previous studies [1], [5]. To reduce the background from sequencing, genes with FPKM less than 1 were removed from analyses. Differentially expressed genes (DEGs) were identified as per the criterion of fold change > 2.0 and p-value < 0.05 (Fig. 2). The identified up-regulated (89) and down-regulated (483) DEGs were used for subsequent bioinformatic analysis, including characterization of the affected hematopoietic genes, perturbed molecular networks and signaling pathways. 75% of zebrafish genes have the homologue in the human genes, and the majority of human genes in zebrafish have the corresponding functions. However, IPA software has not been available for zebrafish gene analysis, therefore, in this study we converted the zebrafish gene to the human gene and analyzed by IPA in order to assess the impacts of Rpl11 deficiency on human DBA pathogenesis using zebrafish model.
Fig. 2.
Differential expressed genes in Rpl11 Mo compared with control Mo.
DEGseq analysis identified 89 significantly up-regulated genes and 483 significantly down-regulated genes in the Rpl11 Mo. The red dots represent up-regulated genes and green dots represent down-regulated genes.
Transcriptome sequencing evaluation
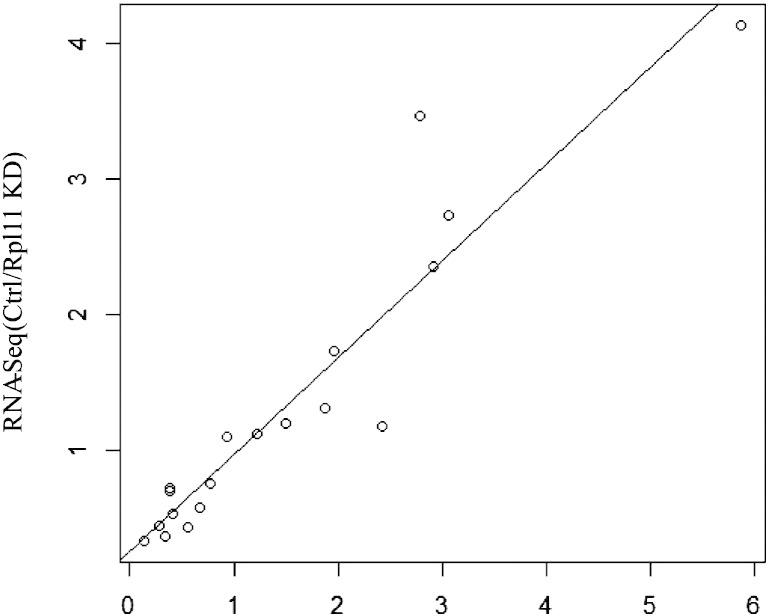
To assess the quality of RNA-Seq sequencing data, we selected some critical genes associated with hematopoiesis and evaluated their gene expression by qPCR analysis. The samples used for qPCR evaluation were from a batch of freshly prepared zebrafish embryo samples with Rpl11 Mo and control Mo. Each qPCR experiment was repeated 3 times to represent the true gene expression level. As shown in Fig. 3, the correlation between RNA-Seq and qPCR results has a good correlation coefficient of 93.7%, indicating that we obtained a representative dataset of transcripts expressed in zebrafish Rpl11 Mo and control embryos.
Fig. 3.
Correlation analysis for RNA-seq and qPCR results.
The relative expression abundances (Ctrl/Rpl11 KD) of 19 genes were selected to represent the results of RNA-seq and qPCR experiments. The correlation coefficient of these two set is 0.937 (p-value = 3.389e − 09).
Conflict of interest
The authors declare that there is no conflict of interest on this study.
Acknowledgments
This research was supported by the National “Twelfth Five-Year” Plan for Science & Technology Support (2013BAI01B09 to H.L. & X.F.), “Strategic Priority Research Program” of the Chinese Academy of Sciences, Stem Cell and Regenerative Medicine Research (XDA01040405), National Natural Science Foundation of China (31371300 to Z.Z., 31100924 to Y.L.), National Key Scientific Instrument and Equipment Development Projects of China (2011YQ03013404 to X.F.), State Key Laboratory of Experimental Hematology Pilot Project Grant (ZK13-05 to Z.Z and ZK12-05 to H.J.).
Contributor Information
Xiaofan Zhu, Email: zhuxf6465@gmail.com.
Xiangdong Fang, Email: fangxd@big.ac.cn.
References
- 1.Aanes H., Winata C.L., Lin C.H., Chen J.P., Srinivasan K.G., Lee S.G., Lim A.Y., Hajan H.S., Collas P., Bourque G. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 2011;21(8):1328–1338. doi: 10.1101/gr.116012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pauli A., Valen E., Lin M.F., Garber M., Vastenhouw N.L., Levin J.Z., Fan L., Sandelin A., Rinn J.L., Regev A. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22(3):577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fullwood M.J., Wei C.-L., Liu E.T., Ruan Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19(4):521–532. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J.Q., Habegger L., Noisa P., Szekely A., Qiu C., Hutchison S., Raha D., Egholm M., Lin H., Weissman S. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc. Natl. Acad. Sci. 2010;107(11):5254–5259. doi: 10.1073/pnas.0914114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vesterlund L., Jiao H., Unneberg P., Hovatta O., Kere J. The zebrafish transcriptome during early development. BMC Dev. Biol. 2011;11(1):30. doi: 10.1186/1471-213X-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]