Abstract
Integrating the analysis of the cistrome of a transcription factor by ChIP-Seq with the study of its transcriptional output by microarray or RNA-Seq analysis is a powerful approach to elucidate the genomic functions of a transcription factor. Recently, we employed this approach to determine the mechanism of action by which the nuclear receptor PPARγ elicits its antitumorigenic effects in lung cancer cells upon activation by TZDs (1). Here we describe in detail the design, contents and quality controls for the gene expression and cistrome analyses associated with our study published in Cell Metabolism in 2014.
Keywords: Lung cancer, Gene expression, Microarray, ChIP-Seq, Bioinformatics
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens/NCI-H1993 and NCI-H2347 lung cancer cell lines |
| Sequencer or array type | Sequencer: Applied Biosystems SOLiD 4hq Array: Illumina Human HT12v4.0 Expression Beadchip |
| Data format | Raw sequence data:xsq, (.csfasta and .qual) Mapped sequence data: bam Raw expression data: Illumina Beadstudio tab-delimited file |
| Experimental factors | Transcription factor binding to DNA; gene expression level changes in response to nuclear receptor agonist treatment |
Direct link to deposited data
Deposited data can be found at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE59736.
Experimental design, materials and methods
ChIP of PPARγ
We generated PPARγ-LAP BAC transgenic NCI-H2347 and NCI-H1993 cell lines using a BAC-transgenesis approach [2], [3], [4]. Cells at 80% confluency (~ 1–1.5 × 107) were cross-linked with 1% formaldehyde for 10 min at 37 °C and quenched with 125 mM glycine at room temperature for 5 min. The fixed cells were washed twice with cold PBS, scraped and transferred into 1 ml PBS containing protease inhibitors (Roche). After centrifugation at 700g for 4 min at 4 °C, the cell pellets were resuspended in 100 μl ChIP lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl [pH 8.1] with protease inhibitors) and sonicated at 4 °C with a Bioruptor (Diagenode) (30 s on and 30 s off at highest power for 12 min). The sheared chromatin with a fragment length of ~ 200–600 bp) was centrifuged at 10,000g for 10 min at 4 °C). One hundred microliters of the supernatant was used for ChIP or as input. A 1:10 dilution of the solubilized chromatin in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl 16.7 mM Tris–HCl [pH 8.1]) was incubated at 4 °C overnight with 6 μg/ml of a goat anti-GFP (raised against His-tagged full-length eGFP and affinity-purified with GST-tagged full-length eGFP). Immunoprecipitations were carried out by incubating with 40 μl pre-cleared Protein G Sepharose beads (Amersham Bioscience) for 1 h at 4 °C, followed by five washes for 10 min with 1 ml of the following buffers: Buffer I: 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl [pH 8.1], 150 mM NaCl; Buffer II: 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl [pH 8.1], 500 mM NaCl; Buffer III: 0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris–HCl [pH 8.1]; twice with TE buffer [pH 8.0]. Elution from the beads was performed twice with 100 μl ChIP elution buffer (1% SDS, 0.1 M NaHCO3) at room temperature (RT) for 15 min. Protein–DNA complexes were de-cross-linked by heating at 65 °C in 192 mM NaCl for 16 h. DNA fragments were purified using QiaQuick PCR Purification kit (Qiagen) and eluted into 30 μl H2O according to the manufacturer’s protocol after treatment with RNase A and Proteinase K.
Outline of the computational analyses
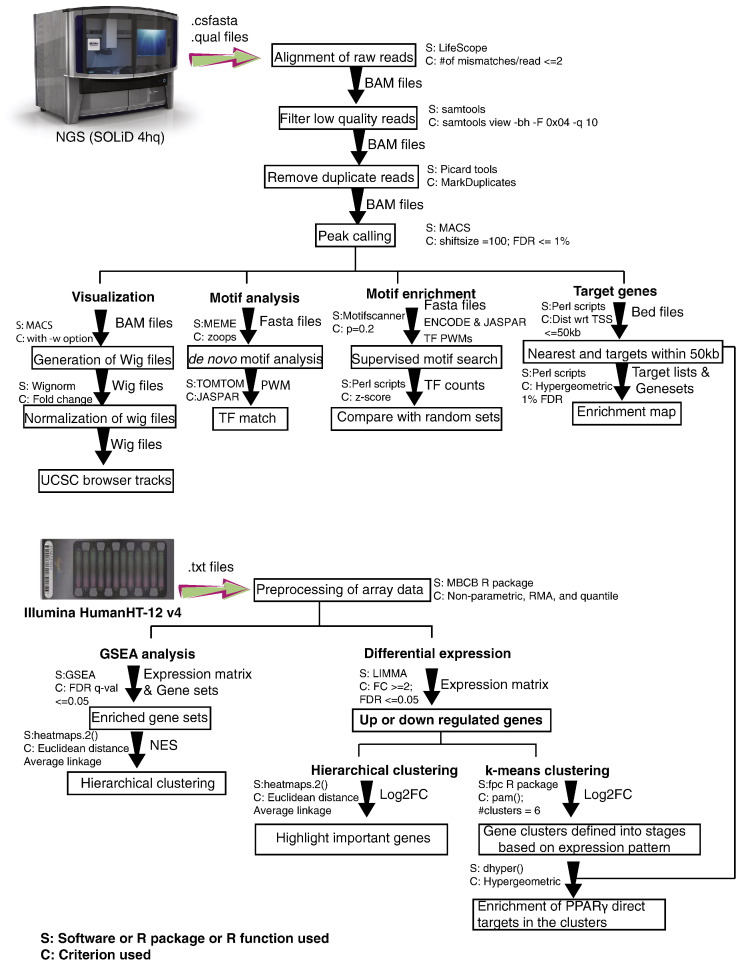
A brief overview of the computational analyses described below is provided in Fig. 1.
Fig. 1.
Flowchart of the computational analyses for this study.
Base calling and alignment
Barcoded libraries of ChIP and input DNA were generated with the SOLiD Fragment Library Barcoding Kit (Applied Biosystems), and 35-nt single-end reads were generated with the SOLiD 4hq system (Applied Biosystems). The images acquired during sequencing runs contain nucleotide information and were translated into DNA sequence, aligned to human reference genome (hg19) using the ChIP-seq module in the LifeScope software. A maximum of two mismatches were allowed during the read alignment. Low-quality reads and duplicate reads were removed from aligned files using “samtools view-bh-F 0 × 04 -q 10” [5] and “Picard MarkDuplicates.jar” commands.
Quality control of next-generation sequencing data
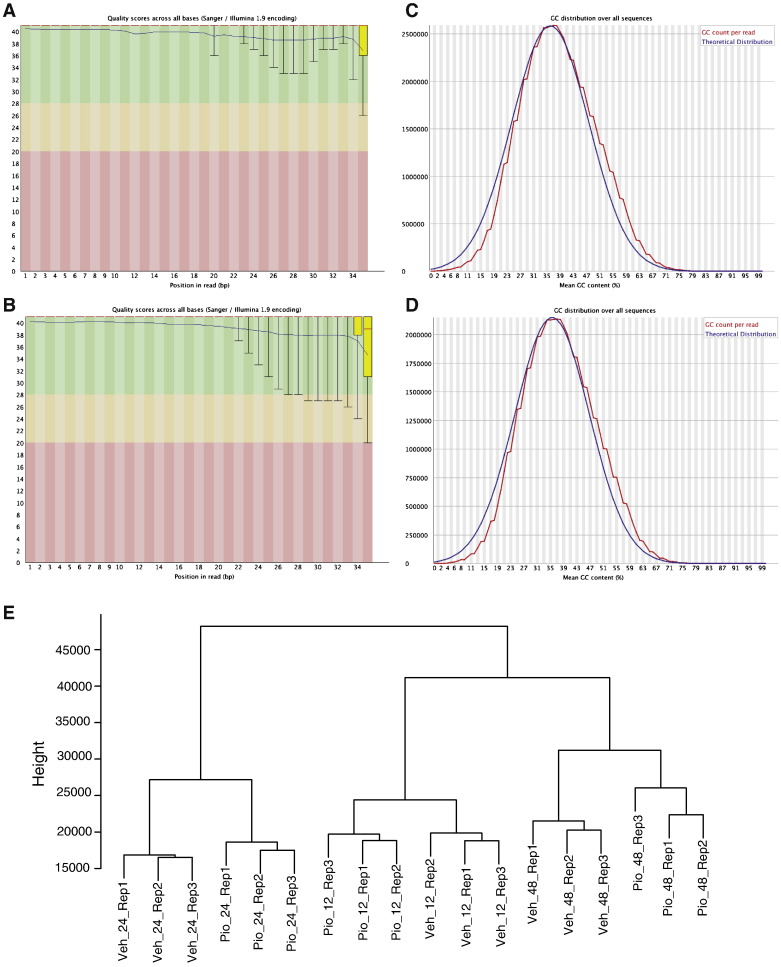
We sequenced all samples twice to ensure sufficient coverage for our ChIP samples. On average, 65% of the reads were mapped to the reference genome. After removing duplicate reads, we performed downstream analyses with 14.04, 25.08, 9.5, and 23.4 million reads for H2347 PPARG ChIP, H2347 input, H1993 PPARG ChIP and H1993 input samples, respectively. Quality control analysis on the mapped reads in H2347 PPARG ChIP and H1993 PPARG ChIP samples using FASTQC indicated no shift in the overall GC content in the sequenced reads and overall excellent quality values (Fig. 2A–D), suggesting that the next-generation sequencing read data are of excellent quality.
Fig. 2.
Quality control analysis for ChIP-seq and gene expression data.
(A and B) Sequencing quality values across all read positions for NCI-H2347 and NCI-H1993 PPARG ChIP sample reads. Yellow box represents the interquartile range. Blue and red lines represent mean and median quality values, respectively. The x-axis depicts each position on the read, the y-axis depicts quality scores.
(C and D) GC content across all read positions for NCI-H2347 and NCI-H1993 PPARG ChIP sample reads. The red curve depicts the GC content distribution per read, the blue curve depicts normal distribution of GC content in the reference genome.
(E) Cluster dendrogram of microarray gene expression profiles. The dendrogram was obtained by hierarchical clustering of pairwise distances between all replicate samples using normalized gene expression values. Sample annotation for Veh_24_Rep1 is vehicle-treated sample, 24 h of treatment, replicate 1 (Pio = pioglitazone-treated samples).
Peak calling
Model-based analysis of ChIP-Seq (MACS) [6] software tool (v.1.4.2) was used to identify PPARγ-bound regions from ChIP-Seq data with 200 bp as the fragment length and read shifting by 100 bp to identify candidate peaks with significant tag enrichment. MACS uses a Poisson distribution model to calculate the significance (p-value) of these peaks. Default parameters of MACS were used for the analyses in our study to quantify the PPARγ ChIP signal fold enrichment over input sample in each region with increased PPARγ occupancy (i.e., peaks), providing this value in the peak file. A false discovery rate (FDR) cutoff of 1% was used to select peaks for further analysis.
Motif discovery and motif enrichment analysis
A crucial step in the secondary and tertiary analyses of our PPARγ ChIP-Seq data was to assess the validity of peak regions identified with MACS. A de novo motif discovery analysis for PPARγ-bound regions shared in NCI-H2347 and NCI-H1993 cells was performed with the Multiple EM for Motif Elicitation (MEME) software tool. MEME uses a multiple sequence alignment approach to identify repeated ungapped sequence patterns in the input DNA with statistical significance [7]. We retrieved 200 bp sequence (i.e., 100 bp sequence flanking the peak summits 3′ and 5′) as input for MEME. From the MEME prediction results, highly enriched motif in terms of number of sites and E-values were selected and then mapped against the transcription factor annotation databases JASPAR [8] and TRANSFAC [9] using TOMTOM suite [10]. TOMTOM identifies transcription factors (TF) position weight matrices (PWMs) also known as motifs similar to the MEME predicted motif. Assuming that the ChIP-Seq data are of high quality, we expect to identify the known motif for the transcription factor with this approach, which is critical to proceed with downstream analyses.
Furthermore, we analyzed the enrichment of known transcription factor motifs in JASPAR and ENCODE by determining the frequency of known motifs in PPARγ-bound regions and in 75,000 random sets of the same sample size by using Motif Scanner [11]. Motif Scanner searches for the known motif instances in the given input sequence over the background model and provides output for the best scoring positions as the motif instances. The background model selected for this study is the 3rd-order Markov model designed using the human promoter sequences in eukaryotic promoter databases (EPD) [11]. The motif enrichment score was calculated as the ratio of the motif frequency in PPARγ binding region set and the mean motif frequency in 75,000 random sets. The Z value and statistical significance (p-value) of the enrichment score was calculated based on the variance and the mean obtained from the 75,000 random simulations.
Target gene analysis and pathway enrichment analysis
Potential protein-coding target genes associated with PPARγ binding regions were identified based on the distance of their transcription start sites (TSSs) (obtained from RefSeq annotation assembly, hg19) to PPARγ binding peak summits. All genes whose TSSs were within 100 kb distance were called as PPARγ target genes. Also, if no gene was identified with in 100 kb distance, the nearest gene was considered as the PPARγ target. Pathway enrichment analysis was performed on the called target genes using gene sets provided by Merico et al. [12]. We performed the hypergeometric test to identify the gene or pathway signatures that were overrepresented in the target gene set. Enrichment p-values were calculated and adjusted for multiple hypotheses testing using the Benjamini–Hochberg method. Significantly enriched pathways (FDR ≤ 1%) were selected and the hypergeometric test was used to calculate the enrichment p-values between each pair of significantly enriched pathways or gene sets. These p-values were used to plot the edges in an enrichment map to represent the strength of enrichment between gene sets in our Cell Metabolism paper [1].
Normalization of wig files
We used the wignorm executable provided in MACS software tools to determine the background signal in the input sample and subtracted it from the ChIP signal. We used the fold change between the ChIP signal and the input signal as the score to build a single wig track to represent the binding strength. This score was used to construct the UCSC browser tracks shown in our Cell Metabolism paper.
Annotation of PPARγ binding regions
We used the annotate Peaks function available in Homer tools [13] to annotate PPARγ binding regions relative to their specific positions in the genome. This function takes the peak coordinates, tag directories as input and extends each tag by their estimated ChIP fragment length, and calculates ChIP fragment coverage represented in per base pair per peak. We used CEAS tool [14] to annotate the binding sites distributed over important genomic features such as promoter, downstream of transcription termination site, untranslated region (UTR), exons and introns.
Microarray data normalization and analyses
For microarray data analyses, NCI-H2347 cells were treated with 50 μM pioglitazone or vehicle (DMSO) for 12, 24 and 48 h, and total DNA-free RNA was prepared. Triplicate experiments were performed for each time point. The whole genome gene expression assay was carried out using Illumina HumanHT-12 v4 Expression BeadChip platform. This array contains more than 47000 probes for the human transcriptome.
Microarray preprocessing is a crucial step to avoid background noise interference with the array experiment results. Hence, the two main steps implemented in our preprocessing of microarray array data were background correction and normalization. In this study, we used a non-parametric version of model-based background correction method (MBCB), which uses an extended model of robust multiarray analysis (RMA), to incorporate the information from negative control beads [15]. These background-corrected data were subjected to quantile normalization to obtain identical sample distributions in terms of their statistical properties.
To assess the differential expression between samples, we used the linear models for microarray data (LIMMA) method [16]. In LIMMA, linear models are fitted to normalized expression data by specifying the design matrix to represent the samples that were used in each array and the contrast matrix to specify which comparisons should be used between RNA samples. Also, LIMMA uses an empirical Bayes method to compute the statistical significance and the fold change between the samples to minimize the standard error. p-values for expression changes were computed and adjusted for multiple hypothesis testing [16]. From the LIMMA results, we selected significant probes as those with significant expression changes between the pioglitazone-treated and vehicle samples at any time point (cutoff: adjusted p < 0.05 and fold change > 2 or < 0.5).
Quality control of gene expression data
We calculated pairwise distances between all array sample expression data using the Manhattan method in dist () R function to check the reproducibility between array sample replicates. This method calculates the absolute distance between the two vectors. We performed hierarchical clustering on this distance matrix using “ward” method in hclust() R function (Fig. 2E).
Clustering analysis
Genes with significant expression change at any given time point were used for clustering analysis. To identify gene clusters that have similar expression patterns, we performed hierarchical clustering with Euclidean distance metric and “agglomerative average” method using heatmap.2 function available in the “gplots” R package [17]. We filtered genes whose expression was not significant and highlighted important genes that are involved in lipid metabolism, oxidative stress and cell cycle regulation in the Cell Metabolism paper [1].
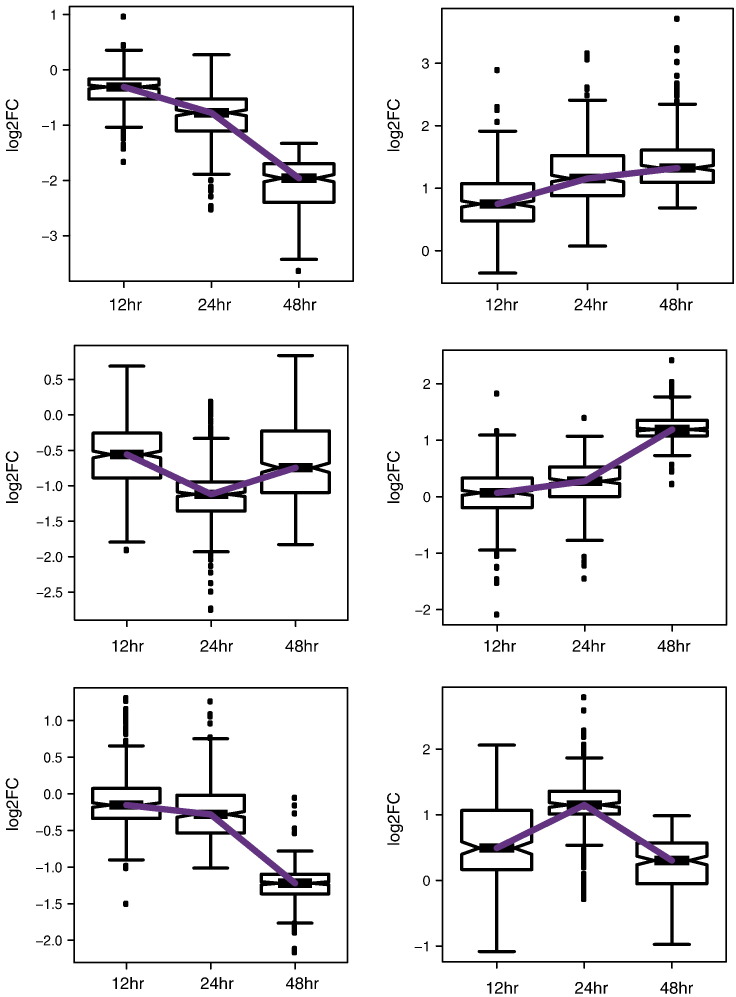
To identify the enrichment of genes are putative direct targets of PPARγ (i.e., genes with PPARγ binding sites near or within 100 kb distance with respect to the TSS, which displayed significant expression changes upon pioglitazone treatment), we clustered genes with significant expression change at any given time point into six clusters using centroid based clustering (k-means clustering) as follows: We calculated the within-group sum of squares measure by increasing the cluster size by one in a stepwise manner. We determined the optimal number of clusters when we did not see a difference in the within-group sum of squares measure after increasing the cluster size. From this analysis, we determined our optimal cluster size as six or eight. We tested the results for both cluster sizes and did not observe new expression patterns when we compared the cluster size of eight to the cluster size of six (data not shown). Hence, we choose the cluster size of six for our analysis. Also, since k-means clustering starts with random initial points and produces different results each time, we used the “pam()” function in the “fpc” R package, which is a more robust method of k-means clustering by starting with defined data points as centers to define clusters. Two of the six clusters showed similar expression pattern between 12, 24, and 48 h time points. Hence, we collapsed them into single cluster (Fig. 3) and quantified the enrichment of PPARγ direct targets in these clusters using the hypergeometric test.
Fig. 3.
Gene expression clusters identified by k-means clustering.
Fold change values for genes that showed significant expression change at any given time point were clustered using k-means clustering, which generated six different clusters. Fold change values for all genes in each cluster at the three time points were plotted to show the expression pattern across different clusters.
Rationale for the use of different distance and linkage methods
For the quality control analysis, we performed hierarchical clustering on all probes on the array, whereas for “significant genes” clustering, we performed hierarchical clustering only for probes of genes with significant expression changes for at least one of the three time points. For the quality control analysis, we used log2-transformed normalized expression values of all probes, whereas for “significant genes” clustering, we used log2-transformed fold change values. The “average method” defines clusters based on the distance between groups, which is calculated as the average of the distances between all pairs of individuals in the groups. Hence, the “average method” is more appropriate to define clusters for differentially expressed genes (i.e., those with significant differences in expression between treated and control samples). Ward’s method defines clusters by minimizing the variance in the distances between two groups. Hence, Ward’s method is more appropriate for analyzing replicate reproducibility. Euclidean distance uses the square root of the sum of the squares of the distances and Manhattan method uses sum of absolute distances. We obtained the same clustering patterns with both methods (data not shown).
GSEA enrichment map
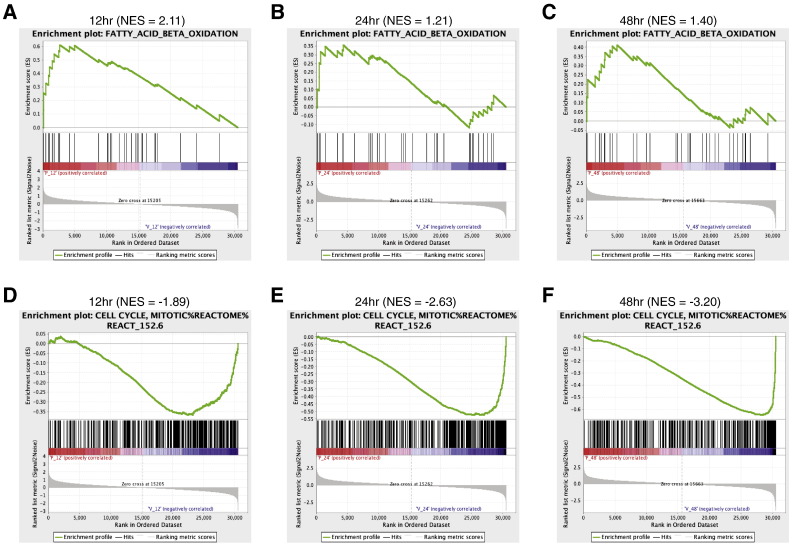
To determine the enrichment of gene sets at the top or bottom of a ranked list of differentially regulated genes upon pioglitazone treatment, we performed gene set enrichment analysis (GSEA) using the time-course gene expression data and a priori defined gene sets. We used default parameters for GSEA except the permutation type parameter. Since, we have a limited number of samples, we used the “geneset” permutation type for the analysis. We performed GSEA for each time point separately (examples for two gene sets are shown in Fig. 4A and B), and the gene sets that were significantly enriched (FDR ≤ 5%) at any given time point were selected for clustering analysis. Hierarchical clustering was performed on the normalized enrichment scores (NES) obtained for all the significant gene sets in all the three time points using heatmap.2 function available in “gplots” R package [17] to produce an enrichment heatmap as displayed in the Cell Metabolism paper [1].
Fig. 4.
GSEA enrichment plots for time-course gene expression data.
(A–C) Enrichment plots for the fatty acid beta-oxidation gene signature.
(D–F) Enrichment plots for the mitotic cell cycle gene signature.
The top part of each plot shows the enrichment score that represents running-sum statistic calculated by “walking down” the ranked list of genes. The middle part shows the position of a member of a gene set in the ranked list of genes. The bottom part depicts the ranking metric that measures a gene’s correlation with a biological function. NES, normalized enrichment score.
Discussion
We describe an integrated analysis of the PPARγ cistrome and gene expression changes caused by the PPARγ agonist pioglitazone in lung cancer cells. This analysis identified the putative direct targets of PPARγ. Multi-step bioinformatic analyses uncovered the gene regulatory program controlled by PPARγ, which provides a hypothesis for its mechanism of action in lung cancer. Importantly, we show that both primary data (for ChIP-seq and gene expression) as well as the primary, secondary and tertiary analysis are of high quality. These data sets and follow-up biochemical studies have been recently used in a study published in Cell Metabolism [1]. Thus, we demonstrate the power of functional genomic approaches coupled with sophisticated bioinformatic analysis to develop mechanistic understanding for the function of transcription factors.
Acknowledgments
This work was supported by the grants RP101251-P06 and RP120732-P3 of the Cancer Prevention and Research Institute of Texas (CPRIT). R.K. is a John L. Roach Scholar in Biomedical Research and a CPRIT Scholar in Cancer Research.
References
- 1.Srivastava N. Inhibition of cancer cell proliferation by PPARgamma is mediated by a metabolic switch that increases reactive oxygen species levels. Cell Metab. 2014;20(4):650–661. doi: 10.1016/j.cmet.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poser I. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods. 2008;5(5):409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hua S., Kittler R., White K.P. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137(7):1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kittler R. A comprehensive nuclear receptor network for breast cancer cells. Cell. Reprogram. 2013;3(2):538–551. doi: 10.1016/j.celrep.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Li H. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey T.L., Williams N., Misleh C., Li W.W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34(Web Server issue):W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32(Database issue):D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wingender E., Dietze P., Karas H., Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24(1):238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S. Quantifying similarity between motifs. Genome Biol. 2007;8(2):R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aerts S. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31(6):1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merico D., Isserlin R., Stueker O., Emili A., Bader G.D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5(11):e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinz S. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin H., Liu T., Manrai A.K., Liu X.S. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25(19):2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 15.Xie Y., Wang X., Story M. Statistical methods of background correction for Illumina BeadArray data. Bioinformatics. 2009;25(6):751–757. doi: 10.1093/bioinformatics/btp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. (Article3) [DOI] [PubMed] [Google Scholar]
- 17.Warnes GR B.B., Bonebakker L., Gentleman R., Liaw W.H.A., Lumley T., Maechler M., Magnusson A., Moeller S., Schwartz M., Venables B. 2012. gplots: Various R Programming Tools for Plotting Data. [Google Scholar]