Abstract
Brackish water lake is the most extraordinary reservoir for bacterial community with an adaptability of tolerance to saline stress. In the present study, metagenomic approach was implemented utilising 454-pyrosequencing platform to gain deeper insights into the bacterial diversity profile of the soil sediment of Chilika Lake, Odisha, India. Metagenome contained 68,150 sequences with 31,896,430 bp and 56.79% G + C content. Metagenome sequences data are now available at NCBI under the Sequence Read Archive (SRA) database with accession no. SRX753382. Bacterial community metagenome sequences were analysed by MG-RAST server representing the presence of 16,212 species belonging to 45 different phyla. The dominating phyla were Proteobacteria, Chloroflexi, Firmicutes, Acidobacteria, Actinobacteria, Bacteroidetes and Planctomycetes. The analysis of bacterial community datasets obtained from two different saline soil sediments revealed significant differences in bacterial community composition and diversity value providing better understanding of the ecosystem dynamics of Chilika Lake.
Keywords: Pyrosequencing, Bacterial diversity, Brackish water, Chilika Lake, Community metagenomics
| Specifications | |
|---|---|
| Organism | Chilika Lake soil sediment metagenome |
| Sex | Not applicable |
| Sequencer or array type | 454 GS junior platform |
| Data format Raw data: | Sff file |
| Experimental factors | Environmental sample |
| Experimental features | 16S rRNA pyrosequencing |
| Analysis using | MG-RAST online server |
| Consent | Not applicable |
| Sample source location | Chilika Lake soil sediment, Odisha, India |
Direct link to deposited data
http://www.ncbi.nlm.nih.gov/sra/SRX753382
Metagenomics provides a valuable tool for discovery of novel genes, metabolic pathways, and important products with biotechnological, pharmaceutical, and medical relevance [1]. High-throughput pyrosequencing of PCR amplicons has emerged as a valuable technique to investigate microbial ecology more thoroughly to unlock the massive uncultured complex microbial diversity present in the environment [2]. Empowerment with these two, in recent times metagenomic strategies have been responsible for exploration of immense microbial diversity worldwide.
Chilika Lake (19° 28′–19° 54′ N; 85° 06′–85° 35′ E) is the largest brackish water lagoon along the Orissa coast, at the east-coast part of India. This lake represents one of the biodiversity hotspots in India and has been identified as a Ramsar site under the convention on ‘Wetlands of international importance’ [3]. The lake covers an average area of 906 km2 in summer and 1065 km2 in rainy season with an average depth of 2 m. It is connected to Bay of Bengal through a zigzag 35 km long outer channel running parallel to the sea [4]. Chilika, is an interesting and unparallel combination of marine, river, and estuarine habitat that supports unique assemblage of marine, brackish water and freshwater microbes [5]. A culture independent study from the sea mouth of Chilika Lake has revealed the substantial phylogenetic diversity of bacterial phyla including Proteobacteria, Spirochaetes, Firmicutes, Chlamydiae, Tenericutes and Planctomycetes [6]. In addition, many novel bacterial species including Shewanella chilikensis sp. nov., a moderately alkaliphilic gammaproteobacterium [7], Streptomyces chilikensis sp. nov., a halophilic streptomycete [8] and Streptomyces barkulensis sp. nov [9] have also been isolated from the Chilika Lake.
In this present work, we have investigated the prokaryotic community structure in the soil sediment of the Chilika Lake at two different salinity gradient by metagenomic approach to find out potential perspective of biotechnological innovations of unexplored lake Chilika. Indeed, the vast majority of microorganisms are yet to be explored in search of biotechnologically precious biomolecules.
Soil sediment samples were collected in triplicate from surface sediments of the Chilika Lake at two different salinity zones, one with moderately high salinity zone (36 PSU) Rambhartia (N1: 19° 39′ 25″N 85° 27′ 43″E) and the other from comparatively less salinity zone (4.66 PSU), Kaluparaghat (N3: 19° 50′ 41″N 85° 24′ 19′E). Metagenomic DNA was extracted by the soil DNA isolation kit (MoBio Laboratories, Carlsbad, CA). Bacterial diversity was analysed by amplification of the V1–V3 region of the 16S rRNA gene according to Basak et al. [10]. Pyrosequencing was performed on a Roche 454 GS-Junior sequencing platform according to the manufacturer's protocol (454 Life Sciences, USA). The pyrosequence output contains 20,140 reads with 10,330,626 bp size (G + C content 56.21%) for sample Rambhartia (N1) and 47,906 reads with 23,265,023 bp size (G + C content 57.03%) for the station Kaluparaghat (N3). Further, the sequences were processed and analysed with MG-RAST on-line server [11].
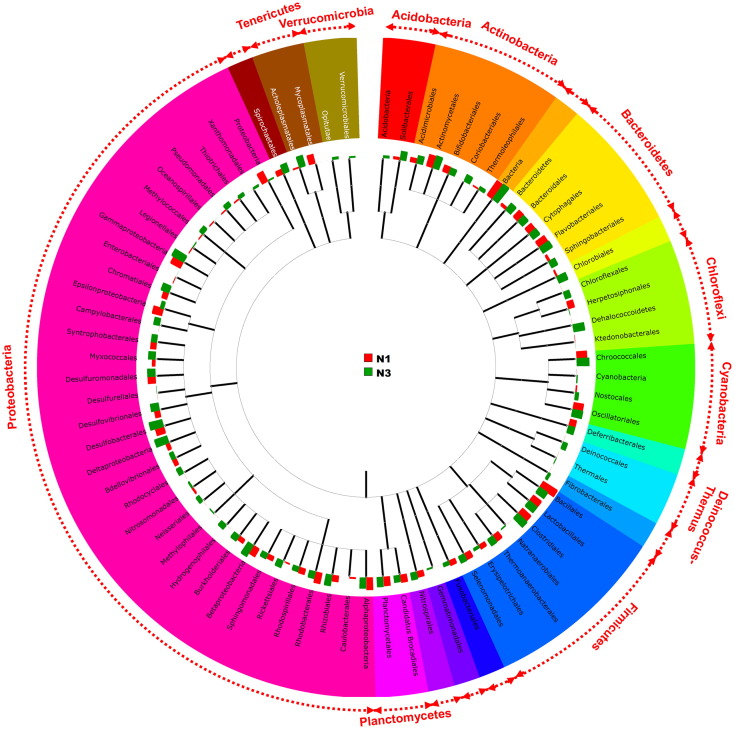
After the analysis of metagenome sequences, a total of 45 phyla were present in the two samples (N1 and N3) of Chilika Lake. In the high salinity zone (Rambhartia) 39 phyla was recorded whereas 44 phyla were observed at low salinity zone (Kaluparaghat) respectively (Fig 1). Interestingly a less number of phyla were observed in the soil sample of Rambhartia, possibly due to the stress produced by high salinity. Proteobacteria is the most dominating phyla in both of the sampling stations. Out of this experimental data, the abundance of Proteobacteria was found to be 35% and 41% at Rambhartia (N1) and Kaluparaghat (N3) respectively. Besides Proteobacteria, Choloroflexi formed 19% and 22% bacterial population in the stations N1 and N3 respectively. The third abundant phyla was Firmicutes at station N1 (15%) but not for the station N3 (1%). Insignificant abundance was detected in both the stations for the phyla Acidobacteria (4%). Most interesting phyla, Actinobacteria accounted 2% for the station at N1 and 3% for N3. Planctomycetes were found to be the less abundant phyla for both stations (N1: 2% and N3: 1%). In summary, the use of 454-pyrosequencing platform allowed us to elucidate the bacterial diversity at two opposite extreme saline zone of Chilika Lake. The rich novel microbial diversity of Chilika Lake, indicates the potentiality to explore new bioactive compounds from the lagoon environment leading immense benefit for industry, therapeutics and basic research.
Fig. 1.
Bacterial community structure of Chilika Lake sediment metagenome.
Nucleotide sequence accession number
Metagenome sequence data from this study were submitted to the NCBI Sequence Read Archive (SRA) under accession numbers: SRR1644053 (Rambhartia: N1), SRR1644054 (Kaluparaghat: N3).
Acknowledgment
This work was financially supported by the NCSCM (National Centre for Sustainable Coastal Management — grant no. 21/RCO/CR/CMR/2013) under the Ministry of Environment and Forests, Government of India. We are indebted to Prof. Dr. Ramesh Ramachandran, Director, NCSCM and Dr. Krishnan P, NCSCM for their encouragement and continuous support. The authors express their sincere thanks to Dr. Ajit Kumar Pattnaik, CDA (Chilika Development Authority) for his support during the cruises and Mr. Niladri Shekher Majumdar, Roche Diagnostics, for his technical support in pyrosequencing work. We are grateful to ICZM project, World Bank for providing the 454 GS Junior pyrosequencer facility for this work.
Contributor Information
Sanghamitra Sengupta, Email: sanghamitrasg@yahoo.com.
Dhrubajyoti Chattopadhyay, Email: djcbcg@caluniv.ac.in.
Maitree Bhattacharyya, Email: bmaitree@gmail.com.
References
- 1.Culligan E.P., Sleator R.D., Marchesi J.R., Hill C. Metagenomics and novel gene discovery. Promise and potential for novel therapeutics. Virulence. 2014;5:399–412. doi: 10.4161/viru.27208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee C.K., Herbold C.W., Polson S.W., Wommack K.E., Williamson S.J., McDonald I.R., Cary S.C. Groundtruthing next-gen sequencing for microbial ecology — biases and errors in community structure estimates from PCR amplicon pyrosequencing. PLoS One. 2012;7:e44224. doi: 10.1371/journal.pone.0044224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahu B.K., Pati P., Panigrahy R.C. Environmental conditions of Chilika Lake during pre and posthydrological intervention: an overview. J. Coast. Conserv. 2014;18:285–297. [Google Scholar]
- 4.Rath J., Adhikary S.P. Distribution of marine macro-algae at different salinity gradients in Chilika lake, east coast of India. Indian J. Mar. Sci. 2005;34:237–241. [Google Scholar]
- 5.Rath J., Adhikary S.P. Biodiversity assessment of algae in Chilika Lake, east coast of India. In: Mohanty P.K., editor. Monitoring and Modelling Lakes and Coastal Environments. Springer; Netherlands: 2008. pp. 22–33. [Google Scholar]
- 6.Parag B., Sasikala Ch., Ramana Ch.V. Molecular and culture dependent characterization of endolithic bacteria in two beach sand samples and description of Rhizobium endolithicum sp. nov. Antonie Van Leeuwenhoek. 2013;104:1235–1244. doi: 10.1007/s10482-013-0046-7. [DOI] [PubMed] [Google Scholar]
- 7.Sucharita K., Sasikala Ch., Park S.C., Baik K.S., Seong C.N., Ramana Ch.V. Shewanella chilikensis sp. nov., a moderately alkaliphilic gammaproteobacterium isolated from a lagoon. Int. J. Syst. Evol. Microbiol. 2009;59:3111–3115. doi: 10.1099/ijs.0.010918-0. [DOI] [PubMed] [Google Scholar]
- 8.Ray L., Suar M., Pattnaik A.K., Raina V. Streptomyces chilikensis sp. nov., a halophilic streptomycete isolated from brackish water sediment. Int. J. Syst. Evol. Microbiol. 2013;63:2757–2764. doi: 10.1099/ijs.0.046284-0. [DOI] [PubMed] [Google Scholar]
- 9.Ray L., Mishra S.R., Panda A.N., Rastogi G., Pattanaik A.K., Adhya T.K., Suar M., Raina V. Streptomyces barkulensis sp. nov., isolated from an estuarine lake. Int. J. Syst. Evol. Microbiol. 2014;64:1365–1372. doi: 10.1099/ijs.0.056614-0. [DOI] [PubMed] [Google Scholar]
- 10.Basak P., Majumder N.S., Nag S., Bhattacharyya A., Roy D., Chakraborty A., SenGupta S., Roy A., Mukherjee A., Pattanayak R., Ghosh A., Chattopadhyay D., Bhattacharyya M. Spatiotemporal analysis of bacterial diversity in sediments of Sundarbans using parallel 16S rRNA gene tag sequencing. Microb. Ecol. 2015;69:500–511. doi: 10.1007/s00248-014-0498-y. [DOI] [PubMed] [Google Scholar]
- 11.Meyer F., Paarmann D., D'Souza M., Olson R., Glass E.M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., Edwards R.A. The metagenomics RAST server — a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinf. 2008;9:1–8. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]