Abstract
This is the first report on the metagenomic approach for unveiling the microbial diversity of Lasundra hot spring, Gujarat State, India. High-throughput sequencing of community DNA was performed on an Ion Torrent PGM platform. Metagenome consisted of 606,867 sequences represent 98,567,305 bps size with an average length of 162 bps and 46% G + C content. Metagenome sequence information is available at EBI under EBI Metagenomic database with accession no. ERP009313. MG-RAST assisted community analysis revealed that 99.21% sequences were bacterial origin, 0.43% was fit to eukaryotes and 0.11% belongs to archaea. A total of 29 bacterial, 20 eukaryotic and 4 archaeal phyla were detected. Abundant genera were Bacillus (86.7%), Geobacillus (2.4%), Paenibacillus (1.0%), Clostridium (0.7%) and Listeria (0.5%), that represent 91.52% in metagenome. In functional analysis, Cluster of Orthologous Group (COG) based annotation revealed that 45.4% was metabolism connected and 19.6% falls in poorly characterized group. Subsystem based annotation approach suggests that the 14.0% was carbohydrates, 7.0% was protein metabolism and 3.0% genes for various stress responses together with the versatile presence of commercially useful traits.
Keywords: Hot spring, Metagenome, Shotgun sequencing, MG-RAST, Microbial diversity, Lasundra
| Specifications | |
|---|---|
| Organism/cell line/tissue | Lasundra hot spring metagenome |
| Sex | Not applicable |
| Sequencer or array type | Ion Torrent PGM platform |
| Data format | Raw data: FASTQ file |
| Experimental factors | Environmental sample |
| Experimental features | Shotgun metagenome sequencing followed by community and functional metagenome analysis using MG-RAST online server |
| Consent | Not applicable |
| Sample source location | Water sample, Lasundra hot spring, Gujarat State, India |
Direct link to deposited data
Deposited data can be found here: https://www.ebi.ac.uk/ena/data/view/PRJNA229814.
The thermal springs are high-temperature aquatic ecosystems and are broadly distributed throughout the world. Microbial life in hot springs is representing a stable and broad scientific interest in the past decades. Thermal hot springs reveal an enormous diversity of microorganisms [1]. The diversity of unculturable bacteria demonstrates a vast gene pool for biotechnological exploration and creates a major face for microbiologists to know their phylogenetic correlation and ecological implication [2]. Besides that metagenomic investigation using high throughput sequencing or library construction shows an enormous consequence for demonstrating microbial composition and functionality in extreme environments [3], [4] and for discovering novel genes [5], [6], [7]. Despite the possibility of the presence of novel microbes with high economic and industrial prospective, there are no detailed reports available on the microbial diversity of most of hot spring of Indian subcontinent. Therefore, the present research was designed to study community and functional microbial diversity with metagenomic approach.
India is one of the most tectonically active areas in the world and has many hot springs. The Lasundra hot spring (N 22°55'30" and E 73°12'33") is one of the less explored sulfuric hot water spring in Kheda District, Gujarat State, India. The temperature of the hot spring reservoir water is reported ranging from 42 °C to 52 °C. A total discharge of water from the spring was about 18,000 l/h, which is used by visitors to take a bath for therapeutic belief. The maximum temperature of the central reservoir was 52 °C and the nearby reservoirs have a temperature varying from 42 °C to 49 °C. Each reservoir was about 12 ft in depth. The base of the reservoir contains stones and algal growth along with soil sediments.
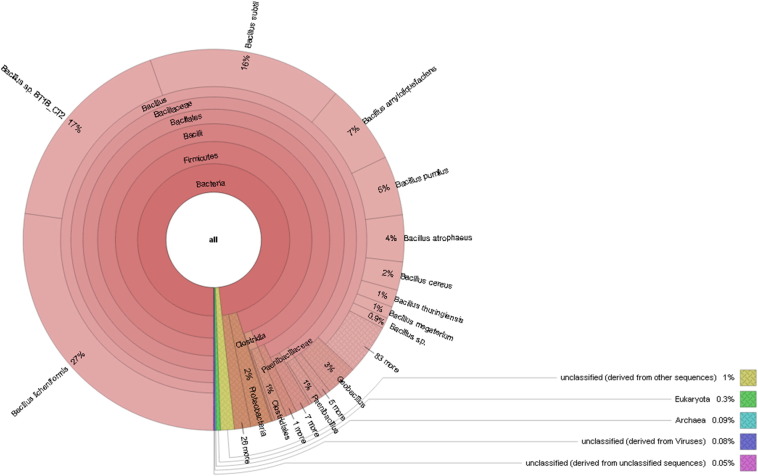
In the present investigation, the total community DNA was isolated from water samples of the spring using a HiPurA™ Water DNA Isolation kit (MB547; Hi-Media, Mumbai, India). Shotgun metagenomic sequencing was performed using the 318 Chip and 300-bp chemistry Ion Torrent PGM platform as per the manufacturer's instructions. Analysis and annotation of output data were carried out by MG-RAST V 3.5 online server using default parameter [8]. The output file contained a total of 606,867 sequences comprised of 98,567,305 bps with 46% G + C content. The community analysis revealed predominance of domain bacteria (99.21%), followed by Eukaryota (0.43%) and archaea (0.11%). A total of 33 prokaryotic phyla including 29 bacterial and 4 archaeal phyla along with 20 eukaryotic phyla were detected. Abundant prokaryotic phyla were Firmicutes (95.5%) followed by Proteobacteria (2.0%), Actinobacteria (0.8%), Bacteroidetes (0.1%), Cyanobacteria (0.1%) and Euryarchaeota (0.09%). At the family level, Bacillaceae (90.1%) followed by Paenibacillaceae (1.3%), Clostridiaceae (0.8%), Listeriaceae (0.5%) and Staphylococcaceae (0.5%) were major dominating bacterial families. Most leading genera were Bacillus (86.7%), Geobacillus (2.4%), Paenibacillus (1.0%), Clostridium (0.7%) and Listeria (0.5%) (Fig. 1).
Fig. 1.
Microbial community structure in Lasundra hot spring metagenome.
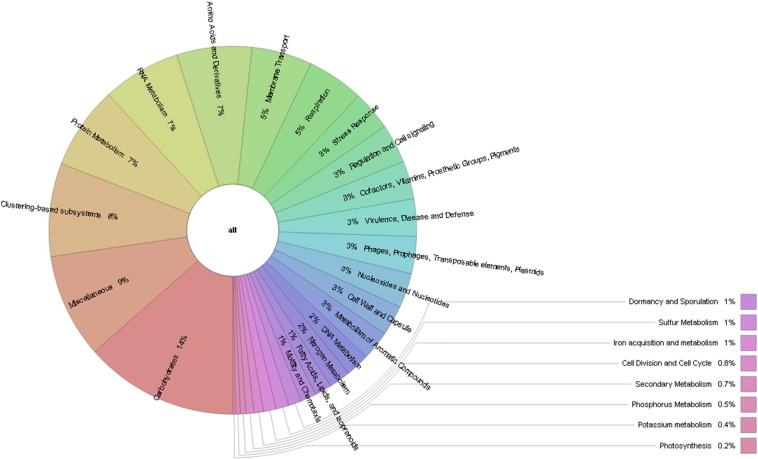
Besides the community analysis, functional analysis was performed with MG-RAST. A total of 388,662 sequences have passed quality control and out of them 76.9% produced 183,408 predicted protein coding regions. From this 183,408 predicted protein features, 66.7% of features assigned an annotation using M5NR database whereas, 33.3% of features have no significant similarities to the protein database. A total of 104,110 features were assigned to functional categories with COG approach and interestingly 45.4% was metabolism connected and 19.6% falls in poorly characterized group that is higher than the earlier metagenome study of saline desert [9] indicating the possibilities of gaining novel gene from the metagenome. Similarly, in subsystem based annotation approach 14.0% was carbohydrates, 7.0% was protein metabolism and 3.0% genes were belonged to stress responses i.e. oxidative stress, periplasmic stress, osmotic stress, heat shock, cold shock, acid stress, and detoxification (Fig. 2). Furthermore, the detection of genes encoded for secondary metabolism and metabolism of aromatic compounds indicates that the hot spring is prosperous with commercially valuable enzymes (Fig. 2). Lipase producing Bacillus licheniformis and Bacillus subtilis were reported from this hot spring earlier, which support our study [10]. The detection of the photosynthetic bacteria, heterotrophs and autotrophs in the hot spring metagenome suggested the nutritive interaction among the microorganism. To the best of our knowledge, this is the first study that describes the complete profiling of microbial diversity from the Lasundra hot spring using the next generation sequencing approach. Metagenomic sequence analysis may provide significant clues in understanding the taxonomic structure and functional potential of Lasundra hot spring with the possibility of getting novel genes and microbes for research and commercial application.
Fig. 2.
Functional structure based on subsystem annotation of Lasundra hot spring metagenome.
Nucleotide sequence accession number
Metagenome sequence data are available on EMBL Metagenomics under the accession no. http://www.ebi.ac.uk/ena/data/view/ERP009313.
Acknowledgment
We are thankful to Ome Research Facility, Anand Agricultural University, Anand, India for providing the facility of the next generation sequencing.
References
- 1.Satyanarayana T., Raghukumar C., Shivaji S. Extremophilic microbes: diversity and perspectives. Curr. Sci. 2005;89(1):78–90. [Google Scholar]
- 2.Sharma R., Ranjan R., Kapardar R.K., Grover A. Unculturable bacterial diversity: an untapped resource. Curr. Sci. 2005;89(1):72–77. [Google Scholar]
- 3.Simon C., Wiezer A., Strittmatter A.W., Daniel R. Phylogenetic diversity and metabolic potential revealed in a glacier ice metagenome. Appl. Environ. Microbiol. 2009;75(23):7519–7526. doi: 10.1128/AEM.00946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie W., Wang F., Guo L., Chen Z., Sievert S.M., Meng J., Huang G., Li Y., Yan Q., Wu S., Wang X., Chen S., He G., Xiao X., Xu A. Comparative metagenomics of microbial communities inhabiting deep-sea hydrothermal vent chimneys with contrasting chemistries. ISME J. 2011;5(3):414–426. doi: 10.1038/ismej.2010.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tirawongsaroj P., Sriprang R., Harnpicharnchai P., Thongaram T., Champreda V., Tanapongpipat S., Pootanakit K., Eurwilaichitr L. Novel thermophilic and thermostable lipolytic enzymes from a Thailand hot spring metagenomic library. J. Biotechnol. 2008;133:42–49. doi: 10.1016/j.jbiotec.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 6.Steele H.E., Jaeger J.E., Daniel R., Streit W.R. Advances in recovery of novel biocatalysts from metagenomes. J. Mol. Microbiol. Biotechnol. 2009;16(1–2):25–37. doi: 10.1159/000142892. [DOI] [PubMed] [Google Scholar]
- 7.Jimenez D.J., Montaña J.S., Alvarez D., Baena S. A novel cold active esterase derived from high Andean forest soil metagenome. World J. Microbiol. Biotechnol. 2012;28(1):361–370. doi: 10.1007/s11274-011-0828-x. [DOI] [PubMed] [Google Scholar]
- 8.Meyer F., Paarmann D., D'Souza M., Olson R., Glass E.M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., Edwards R.A. The metagenomics RAST server‐public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel Rajesh, Mevada Vishal, Prajapati Dhaval, Dudhagara Pravin, Koringa Prakash, Joshi C.G. Metagenomic sequence of saline desert microbiota from wild ass sanctuary, Little Rann of Kutch, Gujarat, India. Genomics Data. 2015;3:137–139. doi: 10.1016/j.gdata.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gohel H.R., Ghosh S.K., Bragazna V.J. Production, purification and immobilization of extracellular lipases from thermophilic Bacillus subtilis Xrf11 and Bacillus licheniformis Xrf12 for production of alkyl esters. IJLBPR. 2013;2(3):278–286. [Google Scholar]