Abstract
MicroRNAs (miRNAs) are small non-coding endogenous RNA molecules that play important roles in various biological processes. This study examined microRNA profiles of Laodelphax striatellus using the small RNA libraries derived from virus free (VF) and rice black-streaked dwarf virus (RBSDV) infected (RB) insects. A total of 59 mature miRNAs (46 miRNA families) were identified as conserved insect miRNAs in both VF and RB libraries. Among these conserved miRNAs, 24 were derived from the two arms of 12 miRNA precursors. Nine conserved L. striatellus miRNAs were up-regulated and 12 were down-regulated in response to RBSDV infection. In addition, a total of 20 potential novel miRNA candidates were predicted in the VF and RB libraries. The miRNA transcriptome profiles and the identification of L. striatellus miRNAs differentially expressed in response to RBSDV infection will contribute to future studies to elucidate the complex miRNA-mediated regulatory network activated by pathogen challenge in insect vectors.
Keywords: MicroRNA (miRNA), Laodelphax striatellus, Rice black-streaked dwarf virus (RBSDV), Deep sequencing
Introduction
RNA interference is the most important and conserved pathway used by eukaryotic organisms against microbial invasion. In the past decade, it has been discovered that virus infection results in the generation of different small RNA (sRNA) populations, including virus derived small interfering RNAs (siRNAs), piwi-interacting RNAs (piRNAs), host microRNAs (miRNAs) etc. [1]. miRNAs are an abundant class of endogenous small non-coding RNA molecules about 22 nucleotides (nt) long. miRNAs regulate gene expression at the post-transcriptional level by binding to target genes in a sequence specific manner, resulting in either translational suppression (animals) or degradation of targeted mRNAs (plants) [2], [3]. Most mature miRNAs are 20–24 nt long and are transcribed from primary-miRNA molecules which contain a characteristic stem loop structure [4]. Among the known miRNAs deposited in miRBase (http://www.mirbase.org), some are highly conserved across different species whereas others are species-specific [5].
Both conserved and species-specific miRNAs play important roles in multiple types of biological functions in eukaryotic cells including development, differentiation, metabolism, apoptosis, etc. [3], [6]. Increasing evidence indicates that complex interactions occur between host/vector/pathogen encoded microRNAs and their corresponding target genes which eventually affect pathogenesis. Host cellular miRNA expression can be greatly influenced during viral invasion and replication by the host antiviral response and by changes of cellular environment [7], [8], [9]. Deep-sequencing technologies provide powerful tools for researchers to investigate a quantitative and in-depth study of miRNA transcriptomes. Expression profiles of host miRNAs in response to various viruses, including human immunodeficiency virus [10], Epstein–Barr virus [11], chikungunya virus [12] etc., contribute to a better understanding of miRNA-mediated host-virus interactions.
The ability of arthropod vectors to transmit many plant and animal viruses is an important factor in the epidemiology of these diseases [13]. The small brown planthopper (Laodelphax striatellus; family Delphacidae, order Hemiptera) is a notorious rice pest in temperate regions mainly because it transmits two economically important rice viruses: rice black-streaked dwarf virus (RBSDV) and rice stripe virus (RSV) [14], [15]. These viruses replicate in both rice and the planthopper vector. We have previously described sRNA libraries derived from L. striatellus infected with RSV/RBSDV or virus free (VF) and comprehensively characterized the siRNAs derived from the viruses [16]. We now report a comparison of the VF and RBSDV infected (RB) sRNA libraries to identify conserved and novel miRNAs of L. striatellus. The profile of differentially expressed planthopper miRNAs was also extensively characterized during virus infection.
Results and discussion
Overview of deep sequencing data in L. striatellus
The small RNA libraries derived from virus free and RBSDV infected L. striatellus constructed in our previous work [16] were used to identify differentially expressed miRNAs in this study. After quality control, length filtering (16–27 nt), adaptor trimming and removal of sRNAs that matches to known non-miRNA classes in Rfam and RepBase database, the number of total clean reads was 3,379,094 and 6,654,452 with unique reads of 1,303,655 and 1,077,947 from the VF and RB libraries, respectively. An overview of the deep sequencing results is presented in Table 1. Different types of non-miRNA sRNAs including transfer RNAs (tRNAs), ribosomal RNA (rRNAs), small nucleolar RNAs (snoRNAs) and small nuclear RNAs (snRNAs) were identified based on the result of Rfam database mapping (Fig. S1). Among these non-coding sRNAs, tRNAs were the most abundant in the total reads, accounting for 88.54% and 96.95% in the VF and RB libraries, respectively. In the unique reads, rRNAs were the most abundant class accounting for more than 50% in both libraries (Fig. S1).
Table 1.
Summary of small RNA sequencing.
| VF library |
RB library |
|||
|---|---|---|---|---|
| Total (%) | Unique (%) | Total (%) | Unique (%) | |
| Junk reads | 8061 (0.14%) | 4453 (0.34%) | 7421 (0.11%) | 3714 (0.34%) |
| Adapter & length filter | 1,480,871 (26.18%) | 345,355 (26.49%) | 2,194,352 (32.98%) | 308,040 (28.58%) |
| Rfam | 786,186 (13.9%) | 33,999 (2.61%) | 1,914,328 (28.77%) | 23,880 (2.22%) |
| Repeats | 2943 (0.05%) | 505 (0.04%) | 3943 (0.06%) | 301 (0.03%) |
| Clean reads | 3,379,094 (59.74%) | 919,490 (70.53%) | 2,534,733 (38.09%) | 742,094 (68.84%) |
| Total reads | 5,656,590 (100%) | 1,303,655 (100%) | 6,654,452 (100%) | 1,077,947 (100%) |
VF, virus free; RB, RBSDV infected.
Conserved miRNAs in L. striatellus
Conserved miRNAs were identified by searching for homologous sequences among the mature miRNAs and miRNA precursors of 31 insect species deposited in the miRBase (Release 20.0). 273,178 and 191,184 total reads accounting for 59 mature miRNAs (46 miRNA families) were identified as conserved insect miRNA candidates in the VF and RB libraries of L. striatellus, respectively. A complete list of these conserved miRNAs is provided in Supplementary File 1.
Most of the reported mature miRNAs show arm selection preference, leading to a high abundance of products from one arm and many fewer from the other arm of the precursor [17]. In this study, 24 miRNAs were derived from the two arms of 12 miRNA precursors, whereas 16 miRNAs were only derived from the 5′ (5p) and 19 miRNAs only from the 3′ (3p) of their precursors. The complementary miRNAs (derived from both arms of the precursor) identified in our study provide strong evidence that these miRNAs were precisely produced from the stem-loop structure of the precursor during miRNA biosynthesis [3]. The arm dominance pattern of these 24 miRNAs were further analyzed. Only one miRNA family (lst-miR-2) was found to have similar read counts from both arms of its precursor, whereas five miRNA families (lst-miR-9, lst-miR-10, lst-miR-281, lst-miR-3049 and lst-let-7) were 5p arm dominant and five miRNA families (lst-miR-8, lst-miR-71, lst-miR-210, lst-miR-276 and lst-miR-993) were 3p arm dominant in both VF and RB libraries (Supplementary File 1). Interestingly, 3p arm dominance of lst-miR-1 was found in the RB library, but there was no arm selection preference for lst-miR-1 in the VF library (Supplementary File 1). How the arm selection pattern of lst-miR-1 was affected by RBSDV infection in L. striatellus needs further investigation.
Analysis of miRNA length shows that the conserved miRNAs of L. striatellus were in the range between 20 and 25 nt with a peak of 22 nt (44.07%), followed by 23 and 21 nt miRNAs (Fig. 1A). The length distribution pattern of L. striatellus miRNAs was consistent with the canonical size of miRNAs produced by Dicer processing and the features of mature miRNAs [3], [18]. In Drosophila, 22 nt miRNAs are produced by Dicer-1 (DCR-1) while Dicer-2 (DCR-2) is responsible for the production of 21 nt siRNAs [19]. Surprisingly, the length distribution analyzed in our previous study indicated that 22 nt RBSDV siRNAs were dominant in infected L. striatellus [16]. Thus, we suspected that DCR-2 in L. striatellus may have the ability to generate both 21 nt host miRNA and 22 nt virus-derived siRNA. This will be an intriguing question for future study. Furthermore, all of the conserved miRNAs were ubiquitously expressed and exhibited a wide range of abundance from 5 to over 10,000 read counts (normalized) in both libraries (Fig. 1B). The differences in abundance of miRNAs in L. striatellus may be due to the functional divergence in the conserved miRNA families.
Fig. 1.
Length and abundance distribution of identified conserved miRNAs in L. striatellus.
(A) Length distribution of conserved miRNAs in L. striatellus. (B) Abundance distribution of conserved miRNAs in L. striatellus (normalized reads). VF: virus free; RB: RBSDV infected.
miRNAs with high expression levels in both VF and RB libraries are shown and compared in Fig. 2. Among all of the conserved miRNA candidates, lst-miR-8-3p was the most abundant in both VF and RB libraries, accounting for 26.19% and 27.29%, respectively (Fig. 2). Notably, the three most abundantly expressed miRNAs (lst-miR-8-3p, lst-miR-184-3p and lst-miR-281-5p) accounted for 64.82% and 64.62% of the total numbers in the VF and RB libraries respectively (Fig. 2). miR-8 and miR-184 have also been found to be the most abundant in other insect species, such as Locusta migratoria [20], Bombyx mori [21], Nilaparvata lugens [22], Aedes albopictus and Culex quinquefasciatus [23], indicating their essential roles in insects. Surprisingly, miR-1, which is commonly highly expressed in various insects and plays key roles in muscle growth [24] and cardiogenesis [25], had less than 100 read copies in the sRNA libraries of L. striatellus (Supplementary File 1).
Fig. 2.
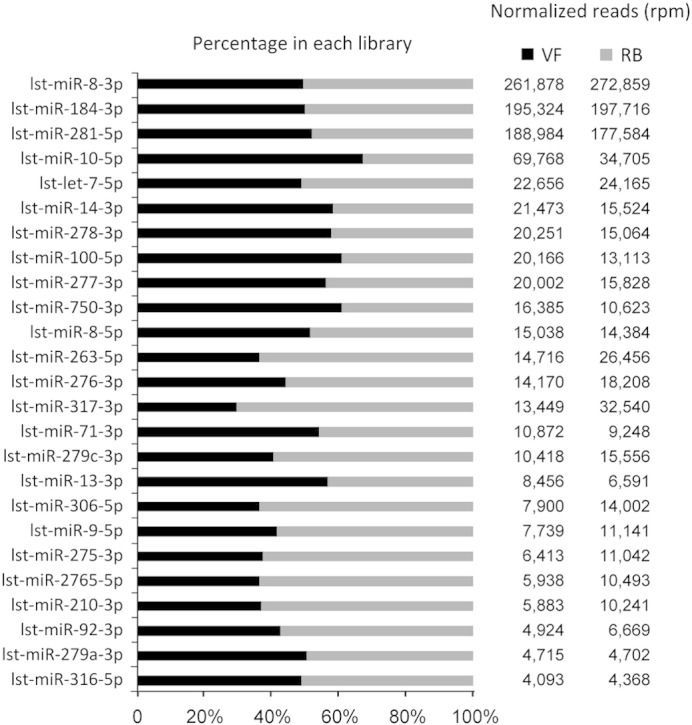
The most abundant conserved miRNAs (normalized reads > 4000) identified in L. striatellus. The left panel represents relative expression levels between VF (black) and RB (gray) libraries. The right panel shows miRNA abundance between VF (black) and RB (gray) libraries with normalized reads. VF: virus free; RB: RBSDV infected.
For a better understanding of the function and evolution of miRNA in insects, it is useful to explore the conservation profile of miRNAs within different insect species [26]. The majority of identified miRNA families in L. striatellus were well conserved among the reported insect miRNAs deposited in miRBase (Supplementary File 2). Among these miRNA families identified in L. striatellus, miR-263 was the most commonly expressed (in 26 out of 31 insect species), whereas miR-996 was only previously identified in Acyrthosiphon pisum (Fig. 3A and Supplementary File 2). Apis mellifera (ame) and Tribolium castaneum (tca) share 45 of their conserved miRNAs with L. striatellus while L. migratoria (lmi) only shares eight as shown in Fig. 3B.
Fig. 3.
Conservation profiles of conserved miRNAs in L. striatellus. (A) Number of insect species identified to have each of the conserved miRNA families identified in L. striatellus. (B) Number of conserved miRNA families in L. striatellus also present in various other insect species. ame: Apis mellifera; tca: Tribolium castaneum; bmo: Bombyx mori; aae: Aedes aegypti; mse: Manduca sexta; api: Acyrthosiphon pisum; hme: Heliconius melpomene; aga: Anopheles gambiae; cqu: Culex quinquefasciatus; nvi: Nasonia vitripennis; dme: Drosophila melanogaster; dan: Drosophila ananassae; dgr: Drosophila grimshawi; dpe: Drosophila persimilis; dwi: Drosophila willistoni; dmo: Drosophila mojavensis; der: Drosophila erecta; dps: Drosophila pseudoobscura; ngi: Nasonia giraulti; dvi: Drosophila virilis; dsi: Drosophila simulans; dya: Drosophila yakuba; nlo: Nasonia longicornis; dse: Drosophila sechellia; lmi: Locusta migratoria.
Differentially expressed conserved miRNAs in response to RBSDV infection
Next generation sequencing provides a powerful approach to investigate pathogen related miRNA profiles and the large number of reads can be used as a reliable source to evaluate the relative abundance of miRNAs between different samples [27], [28]. The expression levels of the conserved miRNAs in the VF and RB libraries were compared to identify miRNAs differentially expressed (more than 1.5-fold change) in response to RBSDV infection. A total of 21 miRNAs were identified, of which nine were up-regulated and 12 down-regulated in RBSDV infected L. striatellus compared to virus free insects (p-value < 0.005) (Fig. 4 and Supplementary File 3). Differentially expressed miRNAs also showed a large variation in abundance ranging from 5 to more than 10,000 (Supplementary File 3). Most miRNA expression differences were in the range from 1.5 to 2 fold. Only three up-regulated (lst-miR-317-3p, lst-miR-1-3p and lst-miR-307-3p) and four down-regulated miRNAs (lst-miR-1-5p, lst-miR-927-5p, lst-miR-932-5p and lst-miR-10-5p) showed differences of greater than 2 fold in response to RBSDV infection (Fig. 4). Significant down-regulation of lst-miR-932-5p was also reported for Aedes aegypti after nine days exposure to Dengue virus type 2 [29], indicating its potential role in response to virus infection. When the shrimp Marsupenaeus japonicus was challenged by a DNA virus, white spot syndrome virus, the expression level of miR-317 was significantly up-regulated [30], which is also similar to the pattern in our studies with L. striatellus and RBSDV. In contrast, miR-927 was significantly over-expressed upon chikungunya virus infection in A. albopictus [12], whereas lst-miR-927-5p was down-regulated in our study. This might be because the up-regulated miR-927 in A. albopictus was from the 3′ arm of the precursor or perhaps because of the complexity of different vector-virus systems. Interestingly, opposite regulation patterns were observed for subset members of miR-1. miR-1-5p was down-regulated in the RBSDV infected L. striatellus sRNA library while miR-1-3p was up-regulated. miR-1 is one of the most conserved miRNAs highly expressed in metazoa and plays important roles in muscle development and heart function regulation in insects [31], [32]. To our knowledge, this is the first report that miR-1 expression levels are affected by pathogen infection in insects. Future investigation is required to explore the importance of these differentially expressed miRNAs in the host antiviral response.
Fig. 4.
Differentially expressed (p-value < 0.005) conserved miRNAs of L. striatellus in response to RBSDV infection. Bars beneath axis x represent down-regulated miRNAs; bars on the upper of axis x represent up-regulated miRNAs.
To validate the reliability of this differential expression of conserved miRNAs of L. striatellus in response to RBSDV infection, qRT-PCR was employed to examine three randomly-selected up-regulated (lst-miR-2765-5p, lst-miR-87-3p and lst-miR-1-3p) and four down-regulated mature miRNAs (lst-miR-750-3p, lst-miR-927-5p, lst-miR-124-3p and lst-miR-133-3p) (Fig. 4). The results with all seven selected miRNAs were consistent with the high-throughput sequencing as expected (Fig. 5) but the fold changes of some miRNAs (such as lst-miR-87-3p, lst-miR-1-3p, lst-miR-750-3p and lst-miR-927-5p) were obviously higher in qRT-PCR than from the sequencing (Fig. 5 and Supplementary File 3). This may be due to different accumulation levels of the virus in the particular insects selected for the samples.
Fig. 5.
qRT-PCR validation of 3 up-regulated and 4 down-regulated conserved miRNAs in response to RBSDV infection. The relative quantification of expression was calculated using the 2− ΔΔCT method. The relative expression levels are presented as the 2− ΔΔCT means ± SE.
Novel miRNA candidates predicted in L. striatellus
Since the full genome of L. striatellus is unavailable, it is difficult to predict potentially novel miRNAs. In our analysis, the transcriptome of L. striatellus and all the ESTs of Delphacidae available in NCBI were used as a genome reference for the prediction of novel miRNA candidates in L. striatellus. A total of 20 novel miRNA candidates were predicted based on the criteria for miRNA identification [33] (Supplementary File 4). These novel miRNA candidates were given names in the form ‘lst-miRn + number’, e.g., lst-miRn3 means ‘L. striatellus miRNA novel number 3’. The negative folding free energy for the predicted novel miRNAs precursor in L. striatellus ranging from − 72.7 kcal/mol to − 19 kcal/mol with an average value of − 32.86 kcal/mol, which is similar to that of Drosophila melanogaster (− 32 kcal/mol) described previously [34]. The typical secondary structures of the predicted novel miRNA precursors were predicted by RNAfold and all of the precursors have the intramolecular capacity to fold into hairpin structures (Fig. 6).
Fig. 6.
Putative folding structure of predicted novel miRNA precursors in L. striatellus. The circles indicate RNA nucleotides and the lines between the circles are bonds that form the primary and secondary structures. The black line beside each structure delimits the mature predicted novel miRNA sequence. VF: Predicted novel miRNA only present in the VF library; RB: Predicted novel miRNA only present in the RB library; VF + RB: Predicted novel miRNA present in both VF and RB libraries.
In previous studies, the predicted novel miRNAs are usually expressed at a very low level compared to the conserved miRNAs. One possible reason for this phenomenon is that conserved miRNAs play key roles in various biological processes, such as metabolism, organ differentiation, development etc., and so are commonly expressed at a high level compared to non-conserved ones [35], [36]. Most of the novel miRNAs we predicted in L. striatellus had less than six copies except for lst-miRn3 and lst-miRn20, which is much lower than that of conserved ones (Supplementary File 1, Supplementary File 4). In addition, several novel miRNAs were only present in the VF library (lst-miRn2, lst-miRn4, lst-miRn8, lst-miRn9, lst-miRn11, lst-miRn13, lst-miRn14, lst-miRn18 and lst-miRn19) or RB library (lst-miRn16) with extremely low copy numbers (one or two) (Supplementary File 4). Numerous previous studies indicate that non-conserved miRNAs are usually expressed in development-specific or stress related patterns [21], [22], [37], [38]. The two small RNA libraries constructed in this study were both derived from the adult stage of L. striatellus, so the low abundance of novel miRNAs may be due to their restricted expression and specific roles in adult L. striatellus. In addition, lst-miRn16, which is only expressed in the RB library, may have a specific function related to RBSDV infection. Further investigation is needed to determine if these novel miRNAs are expressed at a higher level in other developmental stage of L. striatellus or are regulated by various other stresses.
Confirmation of conserved and novel miRNAs in L. striatellus
In order to verify the identified conserved miRNAs and predicted novel miRNAs, RT-PCR validation was performed for four randomly selected conserved and four novel miRNAs of L. striatellus. All the selected miRNAs could be amplified and the PCR products were further confirmed by sequencing (Fig. S2), indicating the high accuracy and efficiency of deep sequencing for miRNA discovery, especially for miRNAs with extremely low expression levels that cannot be detected by traditional methods.
In conclusion, virus free and RBSDV infected small RNA libraries of L. striatellus were comprehensively investigated and compared for miRNA analysis in the present study. A number of conserved and potential novel miRNA candidates were predicted which will greatly contribute to a better understanding of miRNA mediated post transcriptional gene expression in L. striatellus. Furthermore, several differentially expressed conserved miRNAs were identified (nine up-regulated and 12 down-regulated) that may play important roles in the response of L. striatellus to RBSDV infection. Further experimental investigations are needed to study the function of these differentially expressed miRNAs in the antiviral strategy of L. striatellus.
Materials and methods
Insect and virus
A population of virus free insects was maintained on susceptible rice (cv. Wuyujing No. 3) at 26 ± 1 °C, with a photoperiod of 16 h light:8 h darkness and 70 ± 10% relative humidity. RBSDV infected rice plants (provided by Shandong Academy of Agricultural Sciences, China) were used as virus source for inoculation. Virus acquisition, collection of virus free and RBSDV infected insects and total RNA extraction were performed as described in the previous study [16].
Pre-processing of sequencing data
The VF library (NCBI Sequence Read Archive database accession number: SRX255768) and RB library (NCBI Sequence Read Archive database accession number: SRX255770) of L. striatellus constructed in our previous report were used in this study [16]. Low quality sequences (such as A, C, G or T ≥ 80%; Ns ≥ 3; A, C only or G, T only) were filtered from the two libraries and the adapter sequence (5′-TGGAATTCTCGGGTGCCAAGG-3′) was removed using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Clipped high quality sequences were clustered into unique reads and sRNAs with a length of 16–27 nt were identified. The clean sRNA reads were then mapped to Rfam (release 11.0, http://rfam.janelia.org/) and RepBase database (http://www.girinst.org/repbase/) to remove non-coding RNAs (rRNA, tRNA, snoRNA, snRNA) and repetitive RNAs.
Identification of conserved and novel miRNAs
The pre-processed sRNA sequences were further analyzed to identify conserved and novel miRNAs using a proprietary software package, ACGT101-miR v4.2 (LC Sciences, Huston, USA). Mature miRNA and pre-miRNA sequences of all insects deposited in miRBase (release 20, http://www.mirbase.org/) [5] were used as a miRNA reference. Sequences with perfect matches or one mismatch to known insect miRNAs were considered to be conserved miRNAs. miRNAs with low copy reads (fewer than five) were removed during the analysis.
Since the genome sequence of L. striatellus is not available yet, the transcriptome of L. striatellus (NCBI Sequence Read Archive database accession number: SRX016334) together with 138,858 expressed sequence tags (ESTs) of Delphacidae obtained from NCBI were used as a genome reference. For identification of potentially novel miRNAs in L. striatellus, sequences unmapped to insect miRNAs of selected species in miRBase were used as query and mapped to combined EST reference sequences. Based on the mapping results, novel miRNAs were predicted if the extended sequences of the mapped small RNA positions were calculated to form typical hairpin structures and their properties were consistent with the criteria for miRNA identification [33]. The RNAfold webserver was used to predict the stem-loop structure of novel miRNAs (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) with default parameters [39].
Analysis of differential expressed miRNAs
To facilitate comparison of miRNAs in L. striatellus differentially expressed between the VF and RB libraries, read counts from each library were scaled to ‘reads per million’ (rpm) based on the total small RNA read numbers of the corresponding library. A chi-square test using a p-value of < 0.01 was used to identify statistically significant differences between the VF and RB libraries.
miRNA expression profiles by stem-loop quantitative real-time reverse transcription PCR (qRT-PCR)
qRT-PCR was used to confirm the expression profiles of selected miRNAs. A total of 1 μg purified total RNA of L. striatellus (RBSDV-infected and virus free) was used for reverse transcription using an oligo (dT) primer. The stem-loop RT-PCR method was used for validation of differentially expressed miRNAs using an ABI 7500 Real-Time PCR system [40]. The 18S rRNA gene served as an internal control and the 2−△△Ct method was used to evaluate relative expression. The primers used for qRT-PCR are listed in Supplementary File 5. The experiment was done using three independent biological replicates.
Confirmation of conserved and novel miRNAs by RT-PCR and sequencing
Total RNA from virus free L. striatellus was extracted for confirmation. The stem-loop RT-PCR method was used to confirm conserved miRNAs that is same as described for miRNA expression profiles confirmation. PCR products were analyzed with 3% agarose gels and the target bands were then purified and ligated to PMD18-T vector for sequencing by Invitrogen. RT-PCR was used to amplify precursors of predicted novel miRNAs from L. striatellus. Primers were designed based on the miRNA precursor sequences (Supplementary File 5). Target bands of the PCR product were sequenced using the same method as described for conserved miRNAs.
The following are the supplementary data related to this article.
Categories of non-coding small RNAs based on Rfam mapping results in the VF and RB libraries of L. striatellus. VF: virus free; RB: RBSDV infected.
Selected conserved and novel miRNAs validated by RT-PCR and sequencing. (A) Validation of 4 conserved miRNAs. Lane 1: lst-miR-1-3p; lane 2: lst-miR-2765-5p; lane 3: lst-miR-927-5p; lane 4: lst-miR-124-3p. (B) Validation of 4 predicted novel miRNA candidates. Lane 1: lst-miRn15; lane 2: lst-miRn11; lane 3: lst-miRn6; lane 4: lst-miRn2.
Conserved miRNAs identified in L. striatellus.
Conservation profile of conserved miRNAs in L. striatellus.
Differentially expressed conserved miRNAs in response to RBSDV infection.
Novel miRNAs identified in L. striatellus.
Primers used for qRT-PCR.
Conflict of interest statement
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us.
We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
We understand that the corresponding author is the sole contact for the editorial process (including editorial manager and direct communications with the office). He/she is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs. We confirm that we have provided a current, correct email address which is accessible by the corresponding author and which has been configured to accept email from jpchen2001@126.com.
Acknowledgments
This work was financially supported by the State Basic Research Program of China (2014CB138403, 2012CB722504), the International Science & Technology Cooperation Program of China (2012DFA30900), the Special Fund for Agro-scientific Research in the Public Interest of China (201003031) and the Program for Zhejiang Leading Team of Science and Technology Innovation (2009R50032). We thank Professor M.J. Adams, Rothamsted Research, Harpenden, UK for the help in correcting the English of the manuscript.
References
- 1.Ding S.W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 2.Carrington J.C., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Winter J., Jung S., Keller S., Gregory R.I., Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushati N., Cohen S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 7.Skalsky R.L., Cullen B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grassmann R., Jeang K.T. The roles of microRNAs in mammalian virus infection. Biochim. Biophys. Acta. 2008;1779:706–711. doi: 10.1016/j.bbagrm.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh Z., Mallick B., Chakrabarti J. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 2009;37:1035–1048. doi: 10.1093/nar/gkn1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houzet L., Yeung M.L., de Lame V., Desai D., Smith S.M., Jeang K.T. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology. 2008;5:118. doi: 10.1186/1742-4690-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marquitz A.R., Mathur A., Chugh P.E., Dittmer D.P., Raab-Traub N. Expression profile of microRNAs in Epstein–Barr virus-infected AGS gastric carcinoma cells. J. Virol. 2014;88:1389–1393. doi: 10.1128/JVI.02662-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shrinet J., Jain S., Jain J., Bhatnagar R.K., Sunil S. Next generation sequencing reveals regulation of distinct Aedes microRNAs during chikungunya virus development. PLoS Negl. Trop. Dis. 2014;8:e2616. doi: 10.1371/journal.pntd.0002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogenhout S.A., Ammar el D., Whitfield A.E., Redinbaugh M.G. Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 2008;46:327–359. doi: 10.1146/annurev.phyto.022508.092135. [DOI] [PubMed] [Google Scholar]
- 14.Shinkai A. Studies on insect transmission of rice virus diseases in Japan. Bull. Natl. Inst. Agric. Sci. C. 1962;14:1–112. [14: 1–12] [Google Scholar]
- 15.Toriyama S. Rice stripe virus: prototype of a new group of viruses that replicate in plants and insects. Microbiol. Sci. 1986;3:347–351. [PubMed] [Google Scholar]
- 16.Li J., Andika I.B., Shen J., Lv Y., Ji Y., Sun L., Chen J. Characterization of rice black-streaked dwarf virus- and rice stripe virus-derived siRNAs in singly and doubly infected insect vector Laodelphax striatellus. PLoS ONE. 2013;8:e66007. doi: 10.1371/journal.pone.0066007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 18.Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 19.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei Y., Chen S., Yang P., Ma Z., Kang L. Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust. Genome Biol. 2009;10:R6. doi: 10.1186/gb-2009-10-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jagadeeswaran G., Zheng Y., Sumathipala N., Jiang H., Arrese E.L., Soulages J.L., Zhang W., Sunkar R. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics. 2010;11:52. doi: 10.1186/1471-2164-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Q., Lu L., Hua H., Zhou F., Lin Y. Characterization and comparative analysis of small RNAs in three small RNA libraries of the brown planthopper (Nilaparvata lugens) PLoS ONE. 2012;7:e32860. doi: 10.1371/journal.pone.0032860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skalsky R.L., Vanlandingham D.L., Scholle F., Higgs S., Cullen B.R. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genomics. 2010;11:119. doi: 10.1186/1471-2164-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sokol N.S., Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon C., Han Z., Olson E.N., Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marco A., Hui J.H., Ronshaugen M., Griffiths-Jones S. Functional shifts in insect microRNA evolution. Genome Biol. Evol. 2010;2:686–696. doi: 10.1093/gbe/evq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linsen S.E., de Wit E., Janssens G., Heater S., Chapman L., Parkin R.K., Fritz B., Wyman S.K., de Bruijn E., Voest E.E., Kuersten S., Tewari M., Cuppen E. Limitations and possibilities of small RNA digital gene expression profiling. Nat. Methods. 2009;6:474–476. doi: 10.1038/nmeth0709-474. [DOI] [PubMed] [Google Scholar]
- 28.McCormick K.P., Willmann M.R., Meyers B.C. Experimental design, preprocessing, normalization and differential expression analysis of small RNA sequencing experiments. Silence. 2011;2:2. doi: 10.1186/1758-907X-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell C.L., Harrison T., Hess A.M., Ebel G.D. MicroRNA levels are modulated in Aedes aegypti after exposure to Dengue-2. Insect Mol. Biol. 2014;23:132–139. doi: 10.1111/imb.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang T., Xu D., Zhang X. Characterization of host microRNAs that respond to DNA virus infection in a crustacean. BMC Genomics. 2012;13:159. doi: 10.1186/1471-2164-13-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen H.T., Frasch M. MicroRNAs in muscle differentiation: lessons from Drosophila and beyond. Curr. Opin. Genet. Dev. 2006;16:533–539. doi: 10.1016/j.gde.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Asgari S. MicroRNA functions in insects. Insect Biochem. Mol. Biol. 2013;43:388–397. doi: 10.1016/j.ibmb.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Matzke M., Ruvkun G., Tuschl T. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonnet E., Wuyts J., Rouze P., Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- 35.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 36.Sunkar R., Jagadeeswaran G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 2008;8:37. doi: 10.1186/1471-2229-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Surridge A.K., Lopez-Gomollon S., Moxon S., Maroja L.S., Rathjen T., Nadeau N.J., Dalmay T., Jiggins C.D. Characterisation and expression of microRNAs in developing wings of the neotropical butterfly Heliconius melpomene. BMC Genomics. 2011;12:62. doi: 10.1186/1471-2164-12-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo W., Wu G., Yan F., Lu Y., Zheng H., Lin L., Chen H., Chen J. Identification of novel Oryza sativa miRNAs in deep sequencing-based small RNA libraries of rice infected with rice stripe virus. PLoS ONE. 2012;7:e46443. doi: 10.1371/journal.pone.0046443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gruber A.R., Lorenz R., Bernhart S.H., Neubock R., Hofacker I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Lao K.Q., Livak K.J., Guegler K.J. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Categories of non-coding small RNAs based on Rfam mapping results in the VF and RB libraries of L. striatellus. VF: virus free; RB: RBSDV infected.
Selected conserved and novel miRNAs validated by RT-PCR and sequencing. (A) Validation of 4 conserved miRNAs. Lane 1: lst-miR-1-3p; lane 2: lst-miR-2765-5p; lane 3: lst-miR-927-5p; lane 4: lst-miR-124-3p. (B) Validation of 4 predicted novel miRNA candidates. Lane 1: lst-miRn15; lane 2: lst-miRn11; lane 3: lst-miRn6; lane 4: lst-miRn2.
Conserved miRNAs identified in L. striatellus.
Conservation profile of conserved miRNAs in L. striatellus.
Differentially expressed conserved miRNAs in response to RBSDV infection.
Novel miRNAs identified in L. striatellus.
Primers used for qRT-PCR.







