Abstract
Actin, an integral component of the cytoskeleton, plays crucial roles in a variety of cell functions, including cell migration, adhesion, polarity and shape change. Studies performed during the last couple of decades have revealed that the actin also exists in the nucleus. However, the function and properties of nuclear actin remained elusive so far. Recently, we showed that an actin tagged with EYFP and fused with a nuclear localization signal (EYFP-NLS-actin) formed visible filamentous (F)-actin bundles in cells. To obtain further details about the individual genes that are affected by the nuclear actin, we have used the microarray analysis to determine the changes in the expression levels of RNAs in HeLa cells as a result of EYFP-NLS-actin expression. Our results suggest that the nuclear actin plays a role in the activation of genes rather than their repression. The data has been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE59799.
Keywords: Actin, Cell nucleus, Chromatin, Transcription
| Specifications | |
|---|---|
| Organism/cell line/tissue | Human/HeLa cell line |
| Sex | female |
| Sequencer or array type | Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Feature Number version) |
| Data format | Raw data |
| Experimental factors | HeLa cells expressing EYFP-actin or EYFP-NLS-actin |
| Experimental features | RNAs prepared from EYFP-actin and EYFP-NLS-actin expressing HeLa cells, which were separately cultured for 48 h, were used for gene expression analysis by RNA microarray assay. |
| Consent | n/a |
| Sample source location | n/a |
Direct link to deposited data
Experimental design, materials and methods
Plasmids
EYFP-actin and EYFP-NLS-actin expression plasmids were kindly provided by Primal de Lanerolle (University of Illinois, Chicago). EYFP tagged actin behaves like an endogenous actin and has been used to visualize actin dynamics in the inner cells [1]. We have confirmed that the expressed EYFP-NLS-actin accumulates in the nucleus and forms nuclear F-actin bundles [2].
Cell culture and RNA isolation
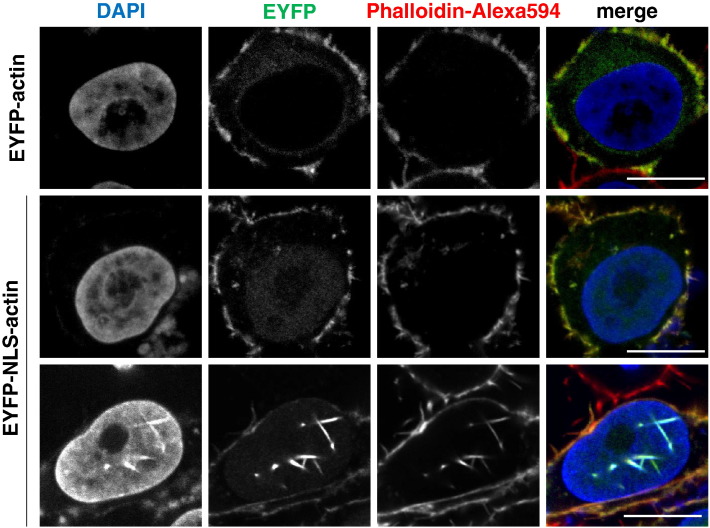
HeLa cells were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum and antibiotics (penicillin and streptomycin) at 37 °C in a 5% CO2 humidified atmosphere. Transfection of HeLa cells was performed using Lipofectamine 2000 (Invitrogen) and 2 μg of plasmid DNA. Captured images of HeLa cells expressing the EYFP-actin or the EYFP-NLS-actin are shown in Fig. 1. Total RNA was isolated from the EYFP-actin or EYFP-NLS-actin expressing HeLa cells by using an RNeasy Mini Kit (QIAGEN) according to the manufacturer's protocol. The quality of isolated RNA was assessed by monitoring the RNA integrity number (RIN). RIN score was calculated by using an Agilent 2100 bio analyzer (Agilent). We confirmed that the RIN scores of the sample RNAs isolated from the EYFP-actin or EYFP-NLS-actin expressing HeLa cell were 10.
Fig. 1.
To analyze the dynamics of actin in the nucleus, an actin fusion protein, in which the actin was fused with the nuclear localization signal (NLS) and EYFP, was ectopically expressed in the HeLa cells (green). Filamentous actin (F-actin) was stained with Phalloidin-Alexa594 (red) and cellular DNA was stained with DAPI (blue). Scale bar = 10 μm.
RNA labeling and hybridization
RNA labeling was performed using a Low Input Quick Amp Labeling Kit (one-color). 0.6 μg of Cyanin-3 (Cy3)-labeled cRNA probe, prepared from 100 ng of total RNA, was fragmented and hybridized to the Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray (GPL16699) using a Gene Expression Hybridization Kit (Agilent) according to the manufacturer's instructions. After hybridization, the array was scanned using an Agilent DNA Microarray Scanner (G2565CA).
Data normalization and comparisons of whole genome expression profiles
The Agilent Feature Extraction software (Agilent) was used to extract raw data from the scanned array images. These raw data files were registered as GEO (GSE59799).
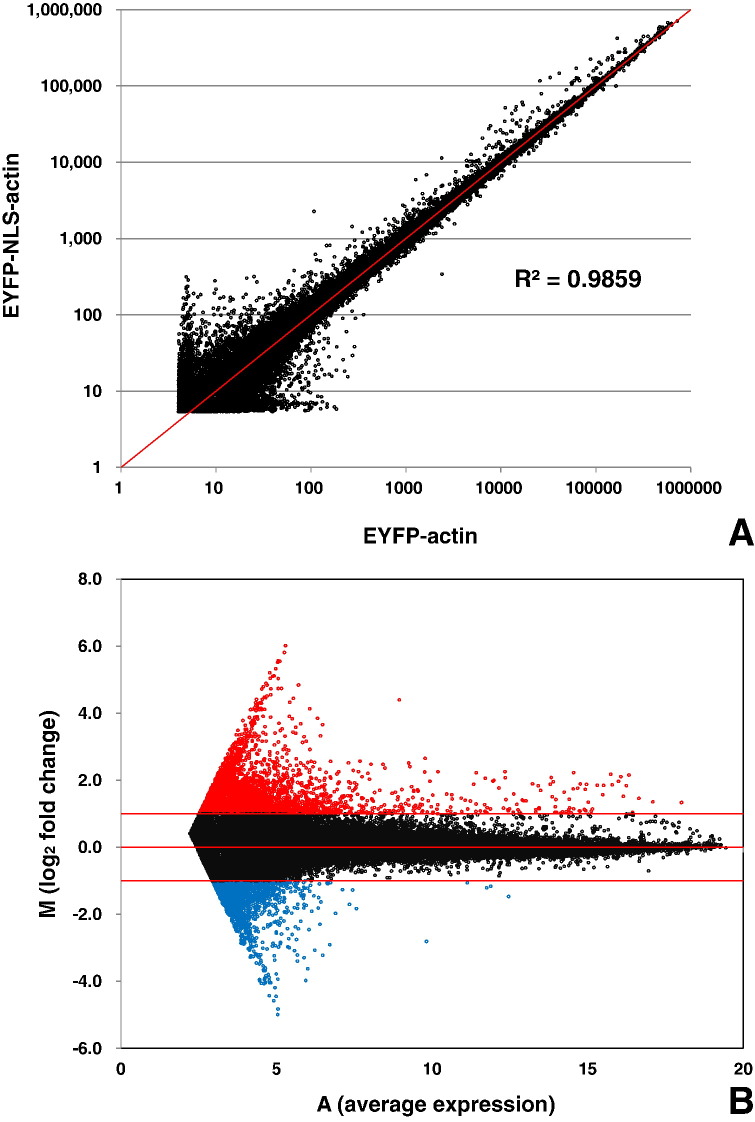
A Scatter plot, showing normalized signal intensity values of different genes in EYFP-actin and EYFP-NLS-actin expressing HeLa cells, is shown in Fig. 2A. As shown, the scatter plot exhibited good correlation coefficient (R = 0.9859) between these signal intensities. This result suggests that the gene expression analysis in these two samples was reproducible. An MA plot is used to identify systematic variations within the arrays, where M is the log ratio (log2 (Exp. gScale Signal) − log2 (Base gScale Signal)) and A is the average log of the product of the two intensities [{log2 (Exp. gScale Signal) + log2 (Base gScale Signal)}/2]. Distribution of up- or down-regulated gene signals is shown Fig. 2B. As shown, the number of up-regulated genes was greater than the number of down-regulated genes, suggesting that the nuclear actin is mainly involved in gene activation rather than in gene repression. This observation is consistent with the results of our previously published reports [2], [3].
Fig. 2.
Scatter plot and MA plot analyses. (A) Scatter plot shows the correlation between the signal intensities of different genes in EYFP-actin and EYFP-NLS-actin expressing HeLa cells. R value is indicated in the plot. (B) MA plot shows the distribution of fold changes in the gene expression. Genes with absolute log fold change > 1 was indicated in red and log fold change <− 1 was indicated in blue.
Conclusion
Herein, we describe the nuclear actin-mediated regulation of gene expression in HeLa cells by using a commercially available RNA microarray. Our microarray analysis data will be useful for elucidating the role of nuclear actin in transcription regulation.
Conflict of interest
Authors declare no conflict of interest.
References
- 1.Herget-Rosenthal S., Hosford M., Kribben A., Atkinson S.J., Sandoval R.M., Molitoris B.A. Characteristics of EYFP-actin and visualization of actin dynamics during ATP depletion and repletion. Am. J. Physiol. Cell Physiol. 2001;281:C1858–C1870. doi: 10.1152/ajpcell.2001.281.6.C1858. [DOI] [PubMed] [Google Scholar]
- 2.Yamazaki S., Yamamoto K., Tokunaga M., Sakata-Sogawa K., Harata M. Nuclear actin activates human transcription factor genes including the OCT4 gene. Biosci. Biotechnol. Biochem. 2015;79:242–246. doi: 10.1080/09168451.2014.972332. [DOI] [PubMed] [Google Scholar]
- 3.Kalendová A., Kalasová I., Yamazaki S., Uličná L., Harata M., Hozák P. Nuclear actin filaments recruit cofilin and actin-related protein 3, and their formation is connected with a mitotic block. Histochem. Cell Biol. 2014;142:139–152. doi: 10.1007/s00418-014-1243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.