Abstract
Very little is known about the genes responsible for Au uptake, reduction and detoxification in plants, which indeed essential to understand the complex trait of AuNP biosynthesis. We designed a targeted experiment to elucidate the response of plant at transcriptional level under Au exposure, and a microarray was performed on root tissue treated with AuCl4− in the absence of nutrient media to record specific gene expression signature. Here, we describe the experimental procedures and data analysis in detail to reproduce the results (available at GEO database under GSE55436) published by Shukla et al. (2014) [1] in the Frontiers in Plant Sciences. The data produced from this study provide significant information of genes which may be used to enhance the AuNP biosynthesis.
Keywords: Au, Arabidopsis, Gene expression, Microarray, AuNPs
| Specifications | |
|---|---|
| Organism/cell line/tissue | Arabidopsis thaliana Col-0 |
| Sex | N/A |
| Sequencer or array type | Affymetrix Arabidopsis Gene 1.0 ST Array |
| Data format | Raw data: CEL files, normalized data: rma-gene-full.chp files, analyzed data: RMALog2_ANOVA Results.csv file |
| Experimental factors | Au treated root tissue vs water treated root tissue |
| Experimental features | Global transcriptome analysis to identify genes involved in AuNP synthesis in Arabidopsis during AuCl4− treatment |
| Consent | N/A |
| Sample source location | Bowling Green, USA (36°59′8″N 86°26′56″W) |
Direct link to deposited data
Experimental design, materials and methods
Experimental scheme
Very few molecular information is available in response to Au treatment, thus we designed this study to get specific gene expression data in response to AuCl4− exposure. Roots of twelve days old hydroponically grown Arabidopsis seedlings were exposed to chloroauric acid (HAuCl4) for 12 h, at pH 4.2 (a natural pH of gold solution), devoid of nutrient media [1]. We preferred the use of hydrogen tetrachloroaurate (HAuCl4) over the potassium tetrachloroaurate (KAuCl4) salt to avoid possibility of unwanted effect of K+ ion, if any.
Plant materials, growth conditions and sampling
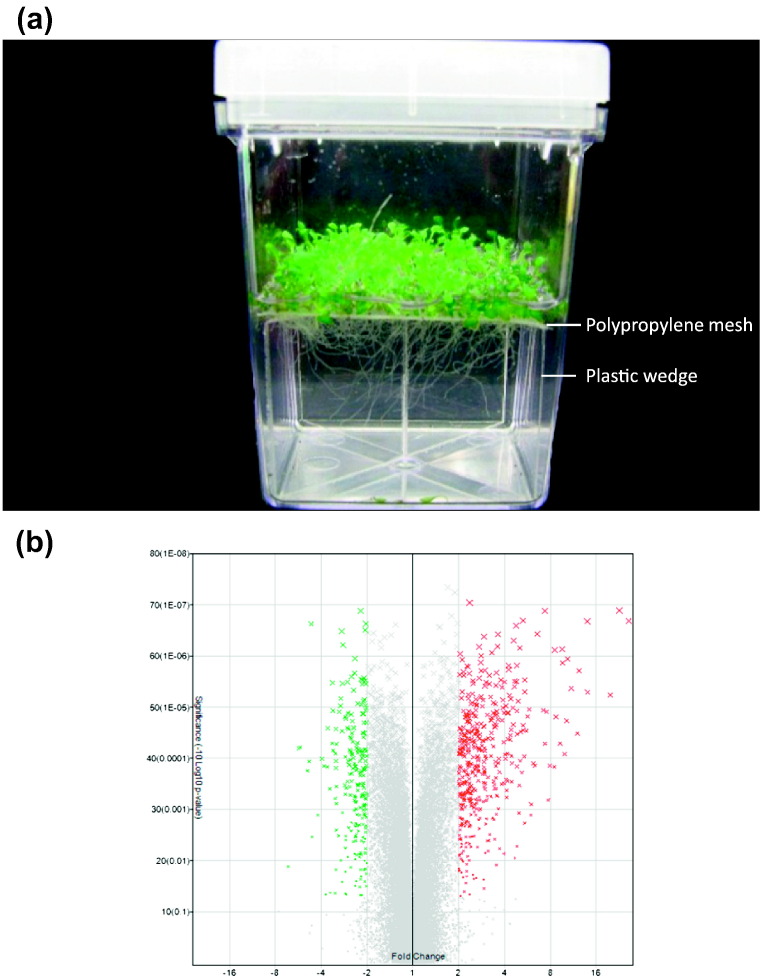
Arabidopsis thaliana seeds ecotype Col-0 procured from Lehle seeds (USA) were surface sterilized using standard procedure and incubated at 4 °C in dark for 48 h for stratification. Approximately 80 seeds were germinated hydroponically [2] on polypropylene mesh (pore sixe 250 μm, Amazon, USA) settled on square plastic wedges in the magenta boxes (GA-7) containing 0.5 × MS medium for 5 days under a long day photoperiod, 16 h cool white fluorescent light (120 μmol m− 2 s− 1)/8 h dark, at 23 °C. Thereafter, seedlings were transferred to 1 × MS medium and allowed to grow for 7 days to get sufficient amount of root tissue (Fig. 1a). After 12 days, the seedlings were transferred into magenta boxes containing 10 ppm AuCl4− (HAuCl4, Sigma-Aldrich, USA) solution at pH 4.2 and incubated for 12 h. Similarly, parallel experiments were performed in distilled water at pH 4.2 to serve as control. After 12 h, root and shoot tissues were separated using sharp blade and snap frozen in liquid nitrogen and stored in − 80 °C until use. To avoid the circadian related effect if any, we performed all operations in the middle of the day. The whole set of experiment repeated three times independently and approximately 240 seedlings were used in each biological replicate.
Fig. 1.
A representative picture of the 12 days old Arabidopsis seedling grown in magenta boxes hydroponically (a). These seedlings treated with 10 ppm of AuCl4− solution or water for 12 h, thereafter total RNA was isolated from root tissues for microarray. Overview of effect of Au exposure on root transcriptome of Arabidopsis (b). Volcano plot shows the significance, and fold change of differentially regulated genes obtained in the present experiment. X axis is the linear fold change calculated for Experiment vs Control; Y axis is − 10log10 p-value of the ANOVA p-values. The gray TCs (transcript clusters) possess < 2-fold change in expression and filtered out. The green TCs show downregulation (> 2-fold) and red TCs show upregulation (> 2-fold).
Total RNA isolation and quality control
Frozen samples were transferred into liquid nitrogen filled polystyrene container to prevent thawing of the tissue during handling. Total RNA was isolated from root and shoot tissue using RNeasy plant Mini Kit equipped with on column DNase I digestion (Qiagen, USA), following the manufacturer's instructions. We ensured the grinding of the root tissues as fine as possible in liquid nitrogen using mortar and pestle as it is the critical factor in isolating high quality RNA from plant tissue. Further, the quality of the total RNA was determined by visualizing the nearly 2:1 ratio of 28S:18S ribosomal RNA by running the samples in the 0.8% TBE (Tris-Borate-EDTA) agarose gel (Supplementary Fig. 1). The concentration and purity of the samples were measured at 260 nm and the ratio of 260 nm/280 nm (generally a ratio ≈ 2, is considered as pure RNA), respectively, using Nanodrop (Wilmington, DE, USA). In addition, quality of the total RNA was also assessed by using 2100 Bioanalyzer (Agilent Technologies) before performing microarray.
Gene expression data analysis
A novel whole-transcript expression array namely “Arabidopsis Gene 1.0 ST Array”, was used to carry out the microarray analysis. Advantage with this system is that it uses the entire transcript to measure expression of a particular gene thereby produces a complete, accurate and unbiased gene expression data. Target preparation, hybridization, washing, staining, and scanning were carried out following the standard protocols provided in the manufacturer's instructions (Affymetrix, USA). Three independent biological experiments were carried out for control and experiment (AuCl4− treated), hence total six Affymetrix array chips were used and no technical replicates were performed. GeneChips were scanned using GeneChip Scanner 3000 7G (Affymetrix) and Affymetrix GeneChip Command Console Software (AGCC version 3.2.4), and six CEL files were generated. Microarray experiments were performed at the Microarray Core Facility, University of Kentucky, USA. All six CEL files were normalized in freely available Expression Console Software v. 1.3.1 (Affymetrix) using RMA algorithm and probe set summarization files (CHP) were generated for genes or exons. The normalized CHP files processed using the freely available secondary analysis tool, Transcriptome Analysis Console software v. 1.0 (Affymetrix), to generate the .csv files containing Differentially Expressed Genes (DEGs) or Exons (DEEs) using a combined criterion of greater than two-fold change with ANOVA p-value of < 0.05 (Condition unpaired) and corrected p-value (FDR) of ≤ 0.16 in the analysis.
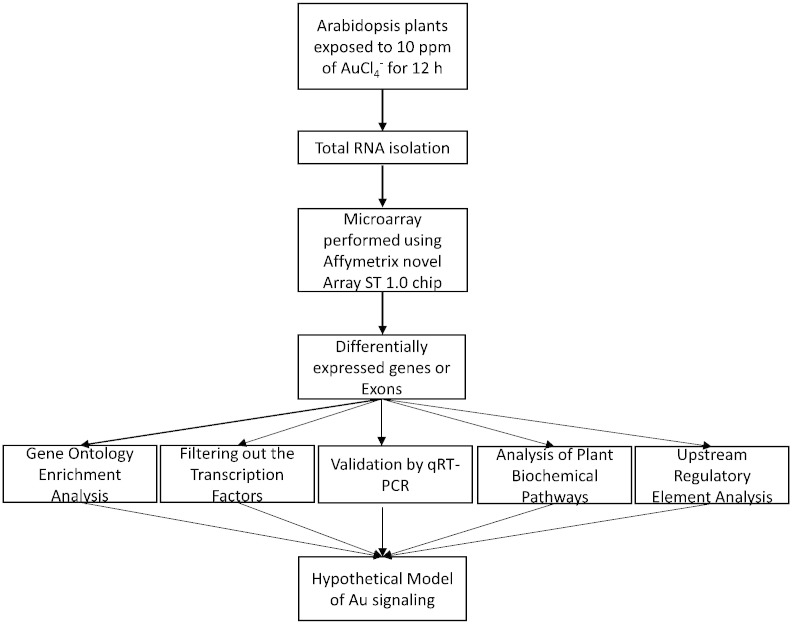
To filter out the known transcription factors from DEGs, we submitted TAIR IDs of DEGs to a public Arabidopsis transcription factor database (AtTFDB) using a web based tool available on this link, http://arabidopsis.med.ohio-state.edu/AtTFDB/ [1]. In order to obtain a view of significantly changed biological function, GO terms affiliated to DEGs were analyzed using singular enrichment analysis (SEA, http://bioinfo.cau.edu.cn/agriGO/index.php) [3] as described in Shukla et al. [1]. Validation of microarray data was carried out by studying the gene expression of 12 key genes using the quantitative RT-PCR method [1]. For performing the quantitative RT-PCR, most of the primers were designed from 5′ or 3′ UTR to produce gene specific results. Effect of Au exposure on biochemical pathways was predicted using the Plant MetGenMap (http://bioinfo.bti.cornell.edu/cgi-bin/MetGenMAP/home.cgi) and Plant Metabolic Network (http://pmn.plantcyc.org/overviewsWeb/celOv.shtml) as described in earlier study [1]. To get the idea of the signaling mechanism operating under Au exposure, we identified overrepresented regulatory element in the 1000 bp upstream sequences of the upregulated genes using the web based Motif finder tool: SCOPE (http://genie.dartmouth.edu/scope/) [4] and the TAIR Motif analysis tool (http://www.arabidopsis.org/tools/bulk/motiffinder/index.jsp) as described in earlier study [1]. We summarized the steps involved in this study and prepared a flow diagram as shown in Fig. 2.
Fig. 2.
Flow diagram of steps involved during this study. Differential gene expression analysis was performed using Expression Console v. 1.3.1 and Transcription Expression Console v. 1.0, software followed by GO analysis, transcription factor identification, biochemical pathway analysis, and enrichment of upstream regulatory element. On the basis of these results, a hypothetical model was proposed showing possible Au mediated signaling in the earlier study [1].
Discussion
Herein, we described a unique dataset of Au transcriptomics in root tissue of Arabidopsis. This dataset is composed of global gene expression measured by using novel unbiased Affymetrix Array ST 1.0. The transcriptomic data is of high quality as evidenced by the RMA analysis of the CEL files. The present dataset has been recently used in a study published in the peer reviewed high impact journal [1], signifying the importance of the data in the context of AuNP synthesis.
The following are the supplementary data related to this article.
Picture shows the samples of total RNA isolated from root tissue of Arabidopsis, analyzed on 0.8% TBE agarose gel for quality control. Samples 1 to 3 represent RNA isolated from Au treated root, and samples 4 to 6 represent RNA isolated from control. Upper bands represent 28S rRNA, lower bands represent 18S rRNA. Intensity ratio of 28S:18S is look like 2:1 which is considered as a universal parameter for quality RNA.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
This research was supported by a grant from the National Science Foundation (Award No. MCB-1158507) to Shivendra V. Sahi.
References
- 1.Shukla D., Krishnamurthy S., Sahi S.V. Genome wide transcriptome analysis reveals ABA mediated response in Arabidopsis during gold (AuCl4−) treatment. Front. Plant Sci. 2014;5:652. doi: 10.3389/fpls.2014.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain A., Sinilal B., Starnes D.L., Sanagala R., Krishnamurthy S., Sahi S.V. Role of Fe-responsive genes in bioreduction and transport of ionic gold to roots of Arabidopsis thaliana during synthesis of gold nanoparticles. Plant Physiol. Biochem. 2014;84:189–196. doi: 10.1016/j.plaphy.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Du Z., Zhou X., Ling Y., Zhang Z., Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq310. (Web Server issue): W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martyanov V., Gross R.H. Using SCOPE to identify potential regulatory motifs in coregulated genes. J. Vis. Exp. 2011;51:e2703. doi: 10.3791/2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Picture shows the samples of total RNA isolated from root tissue of Arabidopsis, analyzed on 0.8% TBE agarose gel for quality control. Samples 1 to 3 represent RNA isolated from Au treated root, and samples 4 to 6 represent RNA isolated from control. Upper bands represent 28S rRNA, lower bands represent 18S rRNA. Intensity ratio of 28S:18S is look like 2:1 which is considered as a universal parameter for quality RNA.