Abstract
Transitions in DNA structure have the capacity to regulate genes, but have been poorly characterised in eukaryotes due to a lack of appropriate techniques. One important example is DNA supercoiling, which can directly regulate transcription initiation, elongation and coordinated expression of neighbouring genes. DNA supercoiling is the over- or under-winding of the DNA double helix, which occurs as a consequence of polymerase activity and is modulated by topoisomerase activity [5]. To map the distribution of DNA supercoiling in nuclei, we developed biotinylated 4,5,8-trimethylpsoralen (bTMP) pull-down to preferentially enrich for under-wound DNA. Here we describe in detail the experimental design, quality controls and analyses associated with the study by Naughton et al. [13] that characterised for the first time the large-scale distribution of DNA supercoiling in human cells (GEO: GSE43488 and GSE43450GSE43488GSE43450).
Keywords: DNA, Supercoiling, Domains, Human, Pull-down
Specifications
| Organism/cell line/tissue | Homo sapiens RPE1 cells |
| Sex | |
| Sequencer or array type | Agilent-028516 Custom human mapping supercoiling array Nimblegen_Human_720K_Chr11extra |
| Data format | Array data was extracted using the RINGO bioconductor packaged and VSN normalised |
| Experimental factors | |
| Experimental features | bTMP pull-down hybridised to microarrays and analysed for DNA supercoil domain distribution |
| Consent | |
| Sample source location |
Direct link to deposited data
Experimental design, materials and methods
Experimental design
Our approach to mapping unrestrained DNA supercoiling utilises the preferential intercalation of TMP into under-wound DNA helices [2], [16]. Using a biotin tagged TMP molecule (bTMP) [14] we are able to enrich for TMP bound DNA by streptavidin pull-down and identify the relative supercoiling across genomic loci by qPCR and microarray analysis. A number of previous studies have compared TMP binding in the presence and absence of transcription and topoisomerase inhibitors to identify transitions in unrestrained DNA supercoiling at promoters and over more extensive regions [1], [2], [5], [7], [9], [10], [11], [12]. However, these studies were unable to adequately compare the distribution of unrestrained DNA supercoiling within samples, as they lack a suitable control for observed TMP bias associated with sequence and chromatin structure [3], [8], [17]. In our experiments we have more thoroughly controlled for the binding and distribution of the bTMP molecule, using a bTMP pull-down of sonicated genomic DNA and in cells treated with the nicking agent bleomycin which relieves DNA supercoils. These bTMP binding controls are essential to understand the relative distribution of DNA supercoiling in vivo as they give a base-line bTMP distribution independent of sequence/chromatin preference. In our experiments bTMP distribution in bleomycin treated cells and genomic DNA are comparable and they can be used interchangeably as the base-level distribution of bTMP binding.
To characterise the distribution of DNA supercoiling in nuclei, the base-line bTMP distribution (log2(bTMP genomic/input)) is subtracted from the bTMP distribution of untreated cells (log2(bTMP control/input)) or cells treated with transcription and/or topoisomerase inhibitors (log2(bTMP inhibitor/input)). In untreated cells this bTMP distribution reflects the steady-state distribution of unrestrained DNA supercoiling in the human genome, with regions of relative under- and over-winding observed over large domains and around gene promoters [13]. Changes in DNA supercoiling after treatment with transcription or topoisomerase inhibitors highlight the dynamic nature of the transitions in DNA structure.
bTMP pull down data
Retinal pigmented epithelial (RPE1) cells with an almost normal human karyotype are incubated in the dark with bTMP (500 μg/ml) followed by 10 minute UV exposure (365 nm) to photo-cross-link bTMP to the DNA in vivo. The DNA is then purified from cells and the incorporation of bTMP confirmed by dotblot using an anti-streptavidin-HRP antibody. A standard ChIP protocol is then used to isolate bTMP bound DNA, followed by whole genome amplification of sample/input DNA and random prime labelling with Cy5 or Cy3 for microarray hybridisation.
We use custom Agilent arrays and Nimblegen 2.1 M promoter arrays according to standard manufacturer's protocols. Microarray samples from Naughton et al. [13] are deposited in GEO under accession numbers GSE43448 and GSE43450GSE43448GSE43450.
Quality control and normalisation
Microarray text files are read, pre-processed and normalised using the RINGO Bioconductor package in R [18]. Arrays are checked for a uniform hybridisation pattern and the signal intensities are compared across arrays to ensure similarity within fluorophore types (Fig. 1). In rare cases arrays show scratches, drying marks and other artefacts that result in them being discarded from subsequent analyses (Fig. 1a). Furthermore, if the observed signal intensity of the pre-normalised arrays has a non-normal distribution for one/both of the fluorophores then this represents a systematic technical bias and these arrays are discarded.
Fig. 1.
Quality control and data normalisation. a) Establishing the array scan quality. Signal distribution across arrays can be used to identify technical problems with array hybridisation including scratches (right hand panel), drying marks, dust particles, etc. b) Intra- and inter-array signal intensity variation is corrected for by VSN normalisation. c) Fluorophore signal intensity bias is corrected for by VSN normalisation. MA plot displaying the observed signal ratio (M) (log2(R)/log2(G)) against the average signal intensity of the two fluorophores (a) ((log(R) + log(G))/2) for a single microarray. R is red and G is green.
In addition to technical problems that prevent the interpretation of microarray data there are inherent biases that can be corrected for through data normalisation [15]. The difference in signal intensity between Cy3 and Cy5 for a single array (intra-array variation) and between individual fluorophores across arrays (inter-array variation) must be accounted for in order to interpret changes in signal intensity between experiments (Fig. 1b pre-normalisation). Additionally, there is a signal intensity bias that is universal to microarray experiments in which the observed signal intensity ratio (M) varies with the average signal intensity ratio (A) (Fig. 1c pre-normalisation). To correct these biases, normalisation is performed using a variance stabilising algorithm (VSN) from the Limma package [4] (Fig. 1b and c post vsn normalisation). Other normalisation procedures, such as a sequential loess and scale normalisation, give almost identical results in the final analyses (data not shown).
To correct for DNA supercoiling independent differences in bTMP binding across array samples, the base-line bTMP bound to genomic DNA is subtracted from bTMP bound in cells. This quantitative measure of bTMP binding allows the comparison of DNA supercoiling across loci in vivo, with positive number showing an enrichment of under-wound DNA and negative numbers showing a depletion of under-wound DNA. All subsequent analysis is performed on the mean of this corrected dataset from duplicate experiments for each experimental condition.
Data analysis
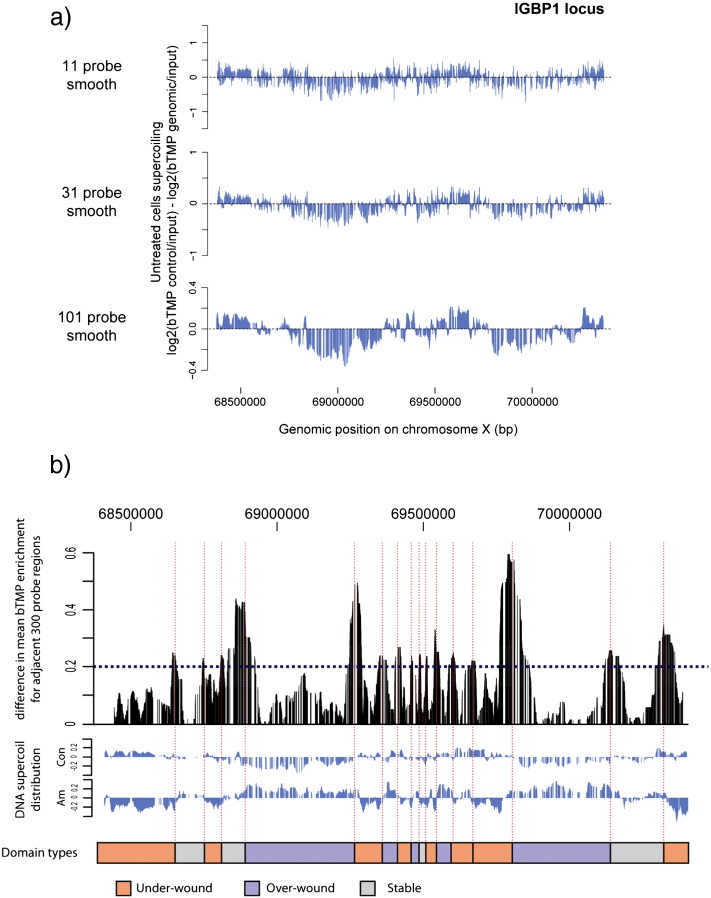
The relative distribution of under-wound DNA in each experimental condition is visualised using the ‘zoo’ package in R [19]. The ‘zoo’ data structure forms an ordered index of observations that is ideal for processing tiled genomic data. Using ‘plot’ to view the ordered distribution of DNA supercoiling across whole loci, domain-scale enrichments and depletions of under-wound DNA are identified. This distribution is clarified by smoothing using the ‘rollingMedian’ function with a window size of 11, 31 and 101 probes (Fig. 2a). DNA supercoil distribution figures are plotted with a 101 probe smoothing (~ 10 kb) to demarcate supercoiling domains [13], but it is vital that subsequent analysis is not performed on heavily smoothed data to avoid a loss of resolution.
Fig. 2.
Identification and classification of DNA supercoil domains. a) Data smoothing demarcates DNA supercoiling domains. Plot of control DNA supercoiling corrected for bTMP binding to genomic DNA smoothed using an 11 probe, 31 probe or 101 probe rolling median. b) An edge filter identifies DNA supercoil domain boundaries. Output of the edge filter (black) identifies peak differences in log2(bTMP control cells/input) − log2(bTMP amanitin treated cells/input) between adjacent 300 probe (~ 30 kb) windows. The cut-off of 0.2 is used to identify major peaks. The boundaries (broken red lines) correspond well with the smoothed control (‘Con’) and amanitin treated (‘Am’) DNA supercoil distributions. Each domain is assigned a type based on whether control is more under-wound, more over-wound or is stable when compared to the alpha amanitin treated DNA supercoil distribution.
To further characterise supercoiling domains observed in the smoothed data a custom edge filter was designed based on Guelen et al. [6]. Smoothed data from bTMP pull-down experiments on untreated cells (‘Con’) and cells treated with the transcription inhibitor α-amanitin (‘Am) identify consistent domain boundaries, with substantial changes to the distribution of DNA supercoiling within these boundaries (Fig. 2b). Therefore, edges were defined based on the difference between ‘Con’ and ‘Am’ DNA supercoil distribution. To define the boundaries a comparison was made for 300 probes up- and down-stream of each probe across a locus, and a cut-off set that matches the most distinct supercoil boundaries (Fig. 2b). To avoid edge effects the 300 probes at the start and end of a locus are removed. In the supercoiling data obtained from Agilent arrays, as presented in Naughton et al. [13], the cut-off was set at 0.2 which captures the most prominent domain boundaries (Fig. 2b). In addition this cut-off identifies a number of very narrow boundaries (< 2000 bp) which appear to correspond to CpG islands (not shown). These boundaries show no relationship to DNA supercoil domains and are removed for subsequent analyses. Therefore, using the edge filter under these conditions 90 domains are identified in 11.9 Mb of Agilent array data and 607 domains are identified across chromosome 11 (135 Mb). Domains are then classified based on the mean change in DNA supercoiling upon transcription inhibition into under-wound (> 0.5), over-wound (< − 0.5) and stable (− 0.5 to 0.5) (Fig. 2b). This classification allows for a direct comparison of regions of the genome with similar DNA supercoil properties in our RPE1 cell-line. Importantly, when repeating these experiments in a new cell line or on a new array platform it is essential to re-calibrate the cut-offs used to define supercoiling domains and their boundaries through careful observation of the underlying data.
Discussion
We describe here a method for mapping DNA supercoiling in vivo through an analysis of bTMP distribution by microarray. By properly controlling for the complex and poorly characterised sequence preference of bTMP using genomic DNA and bleomycin controls, our method can compare the distribution of unrestrained DNA supercoiling within a sample. Using this technique we have identified that ~ 100 kb DNA supercoiling domains exist in human cells, which are modulated by transcription and topoisomerase activity [13]. In future work the bTMP pull-down technique will be used to further probe the distribution and function of DNA supercoiling in genome structure and regulation.
Acknowledgements
This work was funded by the Wellcome Trust 078219/Z/05/Z (N.G.) and Breakthrough Breast Cancer (N.G.). N.G. is a recipient of a UK Medical Research Council senior fellowship (MR/J00913X/1).
References
- 1.Anders L., Guenther M.G., Qi J., Fan Z.P., Marineau J.J., Rahl P.B., Lovén J., Sigova A.A., Smith W.B., Lee T.I. Genome-wide localization of small molecules. Nat. Biotechnol. 2014;32:92–96. doi: 10.1038/nbt.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bermúdez I., García-Martínez J., Pérez-Ortín J.E., Roca J. A method for genome-wide analysis of DNA helical tension by means of psoralen–DNA photobinding. Nucleic Acids Res. 2010;38:e182. doi: 10.1093/nar/gkq687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cech T., Pardue M.L. Cross-linking of DNA with trimethylpsoralen is a probe for chromatin structure. Cell. 1977;11:631–640. doi: 10.1016/0092-8674(77)90080-0. [DOI] [PubMed] [Google Scholar]
- 4.Gentleman R., Irizarry R.A., Carey V.J., Dudoit S., Huber W. Springer Science+Business Media; New York: 2005. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. [Google Scholar]
- 5.Gilbert N., Allan J. Supercoiling in DNA and chromatin. Curr. Opin. Genet. Dev. 2013;25C:15–21. doi: 10.1016/j.gde.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., De Klein A., Wessels L., De Laat W. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 7.Jupe E.R., Sinden R.R., Cartwright I.L. Stably maintained microdomain of localized unrestrained supercoiling at a Drosophila heat shock gene locus. EMBO J. 1993;12:1067–1075. doi: 10.1002/j.1460-2075.1993.tb05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanne D., Straub K., Rapoport H., Hearst J.E. Psoralen–deoxyribonucleic acid photoreaction. Characterization of the monoaddition products from 8-methoxypsoralen and 4,5′8-trimethylpsoralen. Biochemistry. 1982;21:861–871. doi: 10.1021/bi00534a008. [DOI] [PubMed] [Google Scholar]
- 9.Kouzine F., Gupta A., Baranello L., Wojtowicz D., Ben-Aissa K., Liu J., Przytycka T.M., Levens D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ljungman M., Hanawalt P.C. Localized torsional tension in the DNA of human cells. Proc. Natl. Acad. Sci. U. S. A. 1992;89:6055–6059. doi: 10.1073/pnas.89.13.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ljungman M., Hanawalt P.C. Presence of negative torsional tension in the promoter region of the transcriptionally poised dihydrofolate reductase gene in vivo. Nucleic Acids Res. 1995;23:1782–1789. doi: 10.1093/nar/23.10.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto K., Hirose S. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila. J. Cell Sci. 2004;117:3797–3805. doi: 10.1242/jcs.01225. [DOI] [PubMed] [Google Scholar]
- 13.Naughton C., Avlonitis N., Corless S., Prendergast J.G., Mati I.K., Eijk P.P., Cockroft S.L., Bradley M., Ylstra B., Gilbert N. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 2013;20:387–395. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saffran W.A., Welsh J.T., Knobler R.M., Gasparro F.P., Cantor C.R., Edelson R.L. Preparation and characterization of biotinylated psoralen. Nucleic Acids Res. 1988;16:7221–7231. doi: 10.1093/nar/16.15.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siebert M., Lidschreiber M., Hartmann H., Soding J. 2009. A Guideline for ChIP—ChIP Data Quality Control and Normalization (PROT 47) [Google Scholar]
- 16.Sinden R.R., Carlson J.O., Pettijohn D.E. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- 17.Song P.S., Ou C.N. Labeling of nucleic acids with psoralens. Ann. N. Y. Acad. Sci. 1980;346:355–367. doi: 10.1111/j.1749-6632.1980.tb22107.x. [DOI] [PubMed] [Google Scholar]
- 18.Toedling J., Skylar O., Sklyar O., Krueger T., Fischer J.J., Sperling S., Huber W. Ringo—an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinform. 2007;8:221. doi: 10.1186/1471-2105-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeileis A., Grothendieck G. zoo: S3 infrastructure for regular and irregular time series. J. Stat. Softw. 2005;14:1–27. [Google Scholar]