Abstract
A taxonomic description of bacteria was deduced from 5.78 Mb metagenomic sequence retrieved from Tulsi Shyam hot spring, India using bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). Metagenome contained 10,893 16S rDNA sequences that were analyzed by MG-RAST server to generate the comprehensive profile of bacteria. Metagenomic data are available at EBI under EBI Metagenomics database with accession no. ERP009559. Metagenome sequences represented the 98.2% bacteria origin, 1.5% of eukaryotic and 0.3% were unidentified. A total of 16 bacterial phyla demonstrating 97 families and 287 species were revealed in the hot spring metagenome. Most abundant phyla were Firmicutes (65.38%), Proteobacteria (21.21%) and unclassified bacteria (10.69%). Whereas, Peptostreptococcaceae (37.33%), Clostridiaceae (23.36%), and Enterobacteriaceae (16.37%) were highest reported families in metagenome. Ubiquitous species were Clostridium bifermentans (17.47%), Clostridium lituseburense (13.93%) and uncultured bacterium (10.15%). Our data provide new information on hot spring bacteria and shed light on their abundance, diversity, distribution and coexisting organisms.
Keywords: Bacterial diversity, Metagenome, Tulsi Shyam, Hot spring, bTEFAP, Cultivation-independent
| Specifications | |
|---|---|
| Organism/cell line/tissue | Metagenome of Tulsi Shyam hot spring |
| Sex | Not applicable |
| Sequencer or array type | 454 GS FLX pyrosequencing platform |
| Data format | Raw data: FASTQ file |
| Experimental factors | Environmental sample |
| Experimental features | 16S rRNA genes amplified from the metagenome using bTEFAP followed by bacterial community analysis using MG-RAST online server |
| Consent | Not applicable |
| Sample source location | Water sample, Tulsi Shyam hot spring, Gujarat State, India |
Direct link to deposited data
http://www.ebi.ac.uk/ena/data/view/ERP009559.
The temperature in hot springs is usually over the limit of eukaryotic life, which restricts the microbial life to prokaryotes only. In addition to temperature, microbial diversity in hot springs is dictated by environmental physicochemical characteristics including pH, redox potential and concentration of trace elements [1], [2], [3]. Hot springs are well-known sink of industrially interested thermophilic enzymes, encouraging further study to understand the diversity of microbial life in these habitats [4]. Physicochemical properties and geological features of hot springs have been known to geologists for many years. The microbiological study of these ecosystems began in the mid of 20th century [5], and afterward bacterial diversity study of various hot spring were conducted arbitrarily using culture-dependent approach. Though, our understanding regarding the presence and functioning of microbial community in hot spring has been increased drastically with the development of community genomics or metagenomics [4]. Furthermore, high-throughput community sequencing allows quick and inexpensive analysis of microbial diversity in a single run. This leads to revolutionizing microbial ecology studies of extreme environments including saline desert [6], Soda Lake [7] and sulfidic hot springs [8].
Geological Survey of India had identified 340 hot-water springs in the country and classified on the basis of their geo-tectonic setup and grouped into six geothermal provinces. Tulsi Shyam hot spring is arranged under the ‘Deep sedimentary basin of Tertiary age’ situated in Cambay basin in Gujarat state, India [9]. The water temperature of the Tulsi Shyam hot spring is usually between 58 to 67 °C and the pH of the water in the range of 6.8 to 7.7. Hot spring water is clear with no visible turbidity and typical odor of hydrogen sulfide is sensed by visitors. The first microbiological study on isolation of thermostable alkaliphilic bacteria from Tulsi Shyam hot spring was published in 2006 [10]. Subsequently, enzymes producing Anoxybacillus beppuensis [11] and Bacillus amyloliquifaciens [12] bacteria were isolated from the hot spring. To our knowledge, this hot spring has not been investigated so far. The present research is the first study to describe the bacterial diversity present at Tulsi Shyam hot spring. The purpose of this study was to obtain a map of the bacteria population through a culture-independent molecular phylogenetic survey and gain the information on the presence of bacteria valuable for biotechnology.
Water sample was collected from the Tulsi Shyam hot spring (21°3′5″N, 71°1′30″ E), India and subjected to the ultrafiltration using 0.01 μm pore size filter. Retarded material on the filter paper was subjected to extraction of metagenomic DNA using PowerWater® DNA Isolation Kit (MO BIO Laboratories, Inc. CA, USA). Metagenome high-through sequencing has been done by the bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) method [13]. The output was 10,893 sequences with 5,787,661 bp size and 55 ± 1% G + C content. Further, sequences were processed and analyzed with MG-RAST on-line server [14].
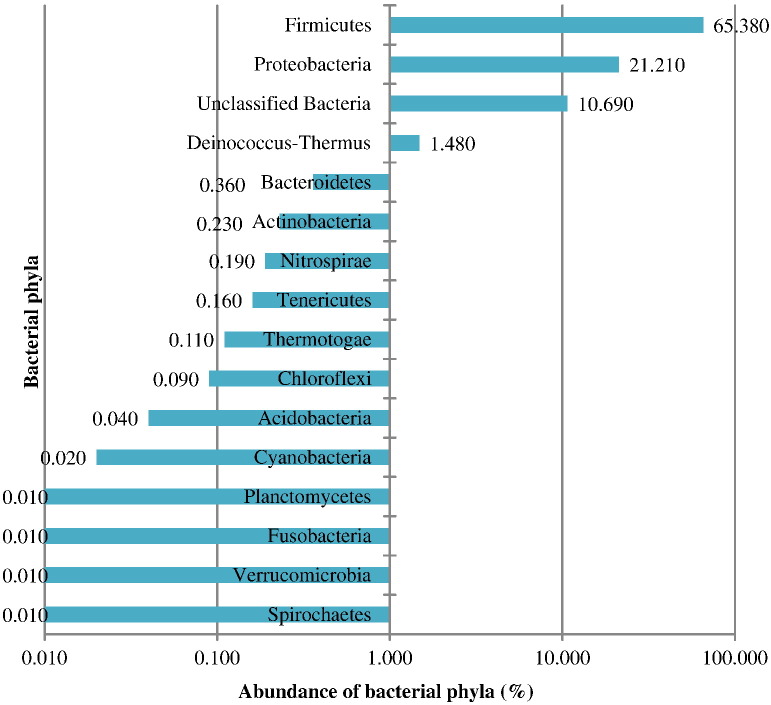
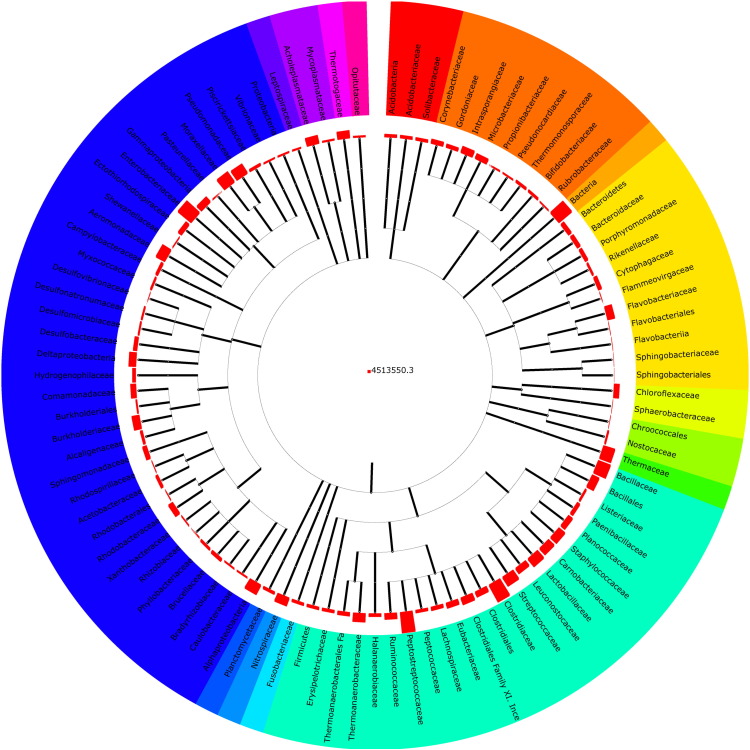
After the analysis of metagenome sequences, a total of 98.2% sequences were belonged to bacteria, 1.5% fits eukaryotic, and 0.3% was unidentified. A total of 16 diverse bacterial phyla including unclassified category were obtained in the metagenome. Prominent phyla were Firmicutes (65.38%), Proteobacteria (21.21%) and unclassified bacteria (10.69%) (Fig. 1). Metagenomes was comprised a total of 97 families with abundant existence of Peptostreptococcaceae (37.33%), Clostridiaceae (23.36%), and Enterobacteriaceae (16.37%) (Fig. 2).
Fig. 1.
Abundance of bacterial Phyla reported in the Tulsi Shyam metagenome.
Fig. 2.
Abundance of bacterial families in the metagenome of Tulsi Shyam hot spring.
At the species level analysis, a total of 287 bacterial species were recognized with the prominent presence of Clostridium bifermentans (17.47%), Clostridium lituseburense (13.93%) and uncultured bacterium (10.15%). Out of 287 species, 15 species that represent 12.34% in metagenome were categorized as an uncultured bacterial species; suggest the hot spring harbor the wealth of uncultivable bacteria. The detection of the human gut bacteria indicates existence of anthropogenic intervention that is due to the exploitation of hot spring reservoir for taking the bath for therapeutic and religious belief. The detection of the thermotolerant and thermophilic species contribute significantly to the composition of the microbial community of hot spring, and whose presence is correlated with the geochemical properties of hot springs. The reported diverse lineages of bacteria help to understand how the physicochemical conditions and biological interactions have shaped these microbial communities within hot spring. The existence of biotechnological significant species in the metagenome suggests the impending application of the hot spring bacteria that evokes the continuing research in this field.
Nucleotide sequence accession number: metagenome sequence data are available on EMBL-Metagenomics under the accession no. ERP009559.
Acknowledgment
Authors are thankful to Research and Testing Laboratory, Lubbock, TX, USA for providing the next generation sequencing facility.
References
- 1.Siering P.L., Clarke J.M., Wilson M.S. Geochemical and biological diversity of acidic, hot springs in Lassen volcanic National Park. Geomicrobiol J. 2006;23:129–141. [Google Scholar]
- 2.Mathur J., Bizzoco R.W., Ellis D.G., Lipson D.A., Poole A.W., Levine R., Kelley S.T. Effects of abiotic factors on the phylogenetic diversity of bacterial communities in acidic thermal springs. Appl. Environ. Microbiol. 2007;73:2612–2623. doi: 10.1128/AEM.02567-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau M.C., Aitchison J.C., Pointing S.B. Bacterial community composition in thermophilic microbial mats from five hot springs in central Tibet. Extremophiles. 2009;13:139–149. doi: 10.1007/s00792-008-0205-3. [DOI] [PubMed] [Google Scholar]
- 4.López-López O., Cerdán M.E., González-Siso M.I. Hot spring metagenomics. Life. 2013;2:308–320. doi: 10.3390/life3020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marsh C.L., Larsen D.H. Characterization of some thermophilic bacteria from the hot springs of Yellowstone National Park. J. Bacteriol. 1953;65:193–197. doi: 10.1128/jb.65.2.193-197.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel R., Mevada V., Prajapati D., Dudhagara P., Koringa P., Joshi C.G. Metagenomic sequence of saline desert microbiota from wild ass sanctuary, Little Rann of Kutch, Gujarat, India. Genomic Data. 2015;2:137–139. doi: 10.1016/j.gdata.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dudhagara P., Ghelani A., Patel R., Chaudhari R., Bhatt S. Bacterial tag encoded FLX titanium amplicon pyrosequencing (bTEFAP) based assessment of prokaryotic diversity in metagenome of Lonar soda lake, India. Genomic Data. 2015;4:8–11. doi: 10.1016/j.gdata.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Headd B., Engel A.S. Biogeographic congruency among bacterial communities from terrestrial sulfidic springs. Front. Microbiol. 2014;5:473. doi: 10.3389/fmicb.2014.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bisht S.S., Das N.N., Tripathy N.K. Indian hot-water springs: a bird's eye view. J. Energy Environ. Carbon Credits. 2011;1:1–15. [Google Scholar]
- 10.Patel R.K., Oza S.M., Rawal C.M. Production and characterization of alkaline protease from thermotolerant alkaliphilic isolates from natural hot spring of Tulshi Shyam, Gujarat. India. Int. J. Biosci. Report. 2006;4:147–151. [Google Scholar]
- 11.Kikani B.A., Singh S.P. The stability and thermodynamic parameters of a very thermostable and calcium-independent α-amylase from a newly isolated bacterium, Anoxybacillus beppuensis TSSC-1. Process Biochem. 2012;47:1791–1798. [Google Scholar]
- 12.Kikani B.A., Singh S.P. Single step purification and characterization of a thermostable and calcium independent α-amylase from Bacillus amyloliquifaciens TSWK1-1 isolated from Tulsi Shyam hot spring reservoir, Gujarat (India) Int. J. Biol. Macromol. 2011;48:676–681. doi: 10.1016/j.ijbiomac.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Dowd S.E., Callaway T.R., Wolcott R.D., Sun Y., McKeehan T., Hagevoort R.G., Edrington T.S. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008;8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer F., Paarmann D., Souza M. D, Olson R., Glass E.M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., Edwards R.A. The metagenomics RAST server — a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinf. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]