Abstract
The ERCC1 and ERCC4 genes encode the two subunits of the ERCC1–XPF nuclease. This enzyme plays an important role in repair of DNA damage and in maintaining genomic stability. ERCC1–XPF nuclease nicks DNA specifically at junctions between double-stranded and single-stranded DNA, when the single-strand is oriented 5′ to 3′ away from a junction. ERCC1–XPF is a core component of nucleotide excision repair and also plays a role in interstrand crosslink repair, some pathways of double-strand break repair by homologous recombination and end-joining, as a backup enzyme in base excision repair, and in telomere length regulation. In many of these activities, ERCC1–XPF complex cleaves the 3′ tails of DNA intermediates in preparation for further processing. ERCC1–XPF interacts with other proteins including XPA, RPA, SLX4 and TRF2 to perform its functions. Disruption of these interactions or direct targeting of ERCC1–XPF to decrease its DNA repair function might be a useful strategy to increase the sensitivity of cancer cells to some DNA damaging agents. Complete deletion of either ERCC1 or ERCC4 is not compatible with viability in mice or humans. However, mutations in the ERCC1 or ERCC4 genes cause a remarkable array of rare inherited human disorders. These include specific forms of xeroderma pigmentosum, Cockayne syndrome, Fanconi anemia, XFE progeria and cerebro-oculo-facio-skeletal syndrome.
Keywords: DNA repair genes, Human, Nucleotide excision repair, Ultraviolet light, Nucleases
1. Introduction
The human ERCC1 and ERCC4 genes encode proteins that associate to form a nuclease that participates in DNA repair and in maintaining chromosome stability. Because the gene products function as partners, they are considered together here. The product of the ERCC4 gene is defective in complementation group F of the inherited human disorder xeroderma pigmentosum (XP-F). Consequently the ERCC4 gene product is commonly referred to as XPF (Lehmann et al., 1994), and that terminology is used in this review. The ERCC1–XPF complex is a two subunit structure-specific nuclease. This nuclease cuts DNA specifically near junctions between single-stranded and double-stranded DNA, where the single strand departs 5′ to 3′ from the junction. ERCC1–XPF plays a central role in nucleotide excision repair (NER) of DNA, a process that removes DNA damage caused by ultraviolet (UV) radiation exposure and by DNA damaging agents that cause covalent helix-distorting adducts (Friedberg et al., 2006; Scharer, 2013). During NER, the ERCC1–XPF nuclease nicks on the 5′ side of the damaged DNA strand, by cleaving an open “bubble” intermediate (Sijbers et al., 1996a; Evans et al., 1997a). ERCC1–XPF also processes 3′ tails of DNA intermediates that occur in some double-strand break repair pathways. Further, ERCC1–XPF participates in repair of DNA interstrand crosslinks. The ERCC1–XPF protein complex is evolutionarily conserved in eukaryotes (Table 1).
Table 1.
Orthologs of the ERCC1 and ERCC4 genes in some commonly studied eukaryotes.
| Human | Mouse | Drosophila | S. cerevisiae | S. pombe |
|---|---|---|---|---|
| ERCC1 | Ercc1 | Ercc1 | RAD10 | swi10 |
| ERCC4 | Ercc4 | Mei-9 | RAD1 | rad16 |
Complete deletion of either ERCC1 or ERCC4 is incompatible with viability in mice or humans. As described below, mutations that impair ERCC4 and ERCC1 function cause several inherited human syndromes associated with increased sensitivity to DNA damage and developmental defects.
2. The ERCC1 and ERCC4 genes
2.1. The ERCC1 gene
The human ERCC1 was the first human DNA repair gene identified by molecular cloning. It was isolated by DNA-based correction of DNA repair-defective Chinese hamster ovary (CHO) cells. Several UV-radiation sensitive CHO cell lines, including lines designated 43-3B (Wood and Burki, 1982) and UV20 (Thompson et al., 1982), were assigned to UV-sensitive genetic complementation group 1 based on cell fusion experiments. Both of these cell lines are defective in forming incisions following UV irradiation (Thompson et al., 1982; Wood et al., 1982). After transfection of the cells with DNA from a human genomic DNA library, subclones could be isolated in which normal sensitivity to UV-radiation had been restored (Westerveld et al., 1984; Rubin et al., 1983). Specific human genomic restriction fragments were identified in such cellular subclones of 43-3B cells (Westerveld et al., 1984). The human gene was designated ERCC1 as an abbreviation for “excision repair cross complementation group 1” because of the cross-species complementation of the human gene with CHO cells. The ERCC1 cDNA was subsequently isolated by utilizing the human DNA present in complemented subclones (van Duin et al., 1986). The last exon of the ERCC1 gene overlaps with the beginning of the CD3E gene, transcribed from the opposite DNA strand (van Duin et al., 1989). CD3E encodes a subunit of the T-cell receptor CD3 complex.
ERCC1 is located on human chromosome at 19q13.32 and comprises 10 exons spanning about 15 kilobases (kb) (van Duin et al., 1987). Four ERCC1 transcript isoforms are annotated, arising from alternative splicing. The canonical transcript encoding functional ERCC1 protein is termed Isoform 1 in the UniProt database (or 202) and is called Isoform 2 in the NCBI Gene database. The other three transcript isoforms encode non-functional transcripts (van Duin et al., 1986). ERCC1 is broadly expressed in mammalian tissues (Wu et al., 2009).
2.2. The ERCC4 gene
The ERCC4 gene was isolated independently by two different strategies. One approach linked several findings. ERCC1 was known to be a homolog of the Rad10 protein from the budding yeast Saccharomyces cerevisiae (van Duin et al., 1986). This was a significant insight because Rad10 had been discovered to associate with yeast Rad1 protein, forming a nuclease involved in NER (Bardwell et al., 1993; Tomkinson et al., 1993). Biochemical experiments showed that ERCC1 functioned together in a complex with ERCC4 (Biggerstaff et al., 1993; van Vuuren et al., 1993). This led to the strong prediction that ERCC4 was a homolog of S. cerevisiae RAD1. Further, it was found that the defect in CHO complementation group 4 cells was identical to the defect in cells from xeroderma pigmentosum group F (XPF) (Biggerstaff et al., 1993; van Vuuren et al., 1993). The cDNA encoding ERCC4 was assembled from fragments homologous to S. cerevisiae Rad1 (Sijbers et al., 1996a). In a second approach, CHO cells in excision repair cross complementing group 4 were used, also defective in incision of DNA following UV radiation (Thompson et al., 1982). One such complementation group 4 cell line, UV41, was used to assign the ERCC4 gene to human chromosome 16p13.1–p13.2 (Liu et al., 1993). The ERCC4 gene was then isolated by complementation of UV41 cells with a human chromosome 16 cosmid library (Thompson et al., 1994). Using the ERCC4 gene sequences, a functional ERCC4 cDNA was assembled (Brookman et al., 1996).
ERCC4 is located on human chromosome 16p13.12 and consists of 11 exons spanning about 28.2 kb (Brookman et al., 1996). In human tissues, the broad tissue expression pattern of ERCC4 is similar to that of ERCC1 (Wu et al., 2009).
3. Biological functions of ERCC1-XPF
3.1. ERCC1-XPF in nucleotide excision repair
Nucleotide excision repair (NER) is a major mechanism for the repair of DNA damaged by chemicals causing helix-distorting covalent adducts and especially for the major UV photoproducts, the cyclobutane pyrimidine dimers and (6-4) photoproducts (Friedberg et al., 2006; Scharer, 2013). ERCC1–XPF is an essential structure-specific endonuclease for the repair of such damaged DNA in both replicating and non-replicating cells. ERCC1–XPF makes incisions on the damaged DNA strand on the 5′ side of the open “bubble” intermediate formed during NER (Sijbers et al., 1996a; Evans et al., 1997a; Mu et al., 1996). In this process (Fig. 1), ERCC1–XPF acts in cooperation with the other protein factors required for the incision process in NER, which are XPC–RAD23B, XPA, RPA, TFIIH and XPG (Aboussekhra et al., 1995).
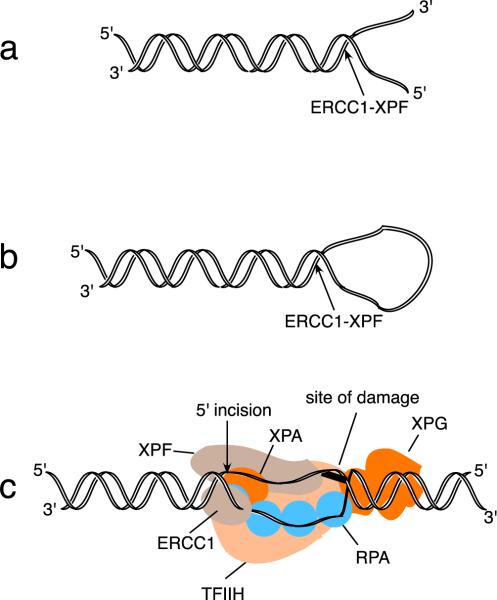
Fig. 1.
Examples of DNA substrates for the ERCC1–XPF nuclease. a. ERCC1–XPF can cleave 3′ ended tails from DNA by nicking within the duplex near the junction between double-stranded and single-stranded DNA. b. ERCC1–XPF can cleave a hairpin structure in DNA with the same polarity as in part a. c. The XPF–ERCC1 nuclease introduces the 5′ incision during NER of an adduct on one strand. Shown here is the open preincision complex, containing TFIIH, XPA, RPA (which includes 4 active DNA-binding oligonucleotide binding (OB) fold domains among its 3 subunits), and XPG. The DNA strands have been separated in an ATP-dependent reaction, which creates a substrate for cleavage by ERCC1–XPF. The cleaved strand departs 5′ to 3′ from the junction, exactly as for the substrates as in parts a and b. The proteins and DNA are shown approximately to scale.
During global genomic nucleotide excision repair (GG-NER), initial distortions in DNA are recognized by XPC-RAD23B (Evans et al., 1997a; Sugasawa et al., 1998). Following this, TFIIH (containing XPB and XPD), XPA, RPA, XPG and ERCC1–XPF converge to form a preincision protein complex, which separates the DNA strands flanking the damaged site. Interactions of ERCC1 with XPA are functionally important within this complex (Volker et al., 2001; Li et al., 1994; Tsodikov et al., 2007; Tripsianes et al., 2007). Transcription-coupled NER (TC-NER) is initiated by the stalling of RNA polymerase II at a lesion on a transcribed DNA strand (Hanawalt, 2002; Hanawalt and Spivak, 2008), after which other repair proteins including CSA, CSB, TFIIH, XPA and RPA are recruited to the damaged site. In both GG-NER and TC-NER, ERCC1–XPF makes an incision on the damaged DNA strand on the 5′ side of the lesion, while XPG makes an incision on the 3′ side of the lesion (O'Donovan et al., 1994).
The 5′ incision and the 3′ incisions occur nearly simultaneously during NER, with uncoupled incisions occurring occasionally on either side (Moggs et al., 1996; Gaillard and Wood, 2001). During the normal course of NER, ERCC1–XPF makes the 5′ incision, a DNA repair polymerase initiates synthesis of a repair patch by copying the undamaged DNA strand, and a 3′ incision is then made by XPG (Staresincic et al., 2009). In human cells, an incision fragment containing the damaged DNA is then released with a modal length of 27–29 nucleotides (Moggs et al., 1996; Bessho et al., 1997a; Huang et al., 1992).
3.2. Double-strand break repair
ERCC1–XPF-deficient cells are moderately more sensitive than normal to double strand break (DSB) inducing agents including ionization radiation produced by x-rays and γ-rays (Wood et al., 1983; Murray et al., 1995; Murray and Rosenberg, 1996; Ahmad et al., 2008). Following exposure to ionizing radiation, ERCC1–XPF-deficient mice and mouse fibroblasts show increased γH2AX foci (a marker of DNA damage), as well as chromosomal abnormalities such as radial structures, gaps and breaks, which are consequences of DSBs (Ahmad et al., 2008). These data indicate that ERCC1–XPF has an important role in one or more pathways of DSB repair. The homologous Rad1–Rad10 nuclease in S. cerevisiae (Ivanov and Haber, 1995; Paques and Haber, 1999) plays a role in the single-strand annealing sub-pathway of homologous recombination. This pathway involves enzymatic processing that yields 3′ DNA overhangs that must be removed to complete repair (Fishman-Lobell and Haber, 1992; Prado and Aguilera, 1995). In mammalian cells, ERCC1 is also required for the removal of long non-homologous ends in targeted homologous recombination (Adair et al., 2000). An ERCC1 defect similarly causes a failure to repair DNA intermediates during single-stranded annealing recombination (Sargent et al., 1997; Al-Minawi et al., 2008, 2009; Bennardo et al., 2008).
Non-homologous end-joining (NHEJ) mediated repair of DSB operates without requiring extensive homology for recombination (Mills et al., 2004). Yeast Rad1–Rad10 processes 3′ ends during microhomology mediated end joining (Ma et al., 2003; McVey and Lee, 2008). Similarly, in mammalian cells, ERCC1–XPF plays a role in removing the 3′ non-homologous ends of some double strand breaks during end-joining. This end joining involving ERCC1–XPF proceeds by an “alternate end-joining” pathway independent of the classical Ku70–Ku80 pathway (Ahmad et al., 2008; Bennardo et al., 2008). Immunoglobulin class-switching proceeds by end-joining pathways, and there is a modest reduction in class-switching in ERCC1-defective mouse B-cells (Schrader et al., 2004).
3.3. Interstrand crosslink repair
DNA interstrand crosslinks (ICLs) are challenging to repair because they obstruct DNA replication and transcription (Klein Douwel et al., 2014; Hodskinson et al., 2014). ERCC1 and XPF-deficient cells are exceptionally sensitive to agents that can form ICLs such as cisplatin, psoralen and mitomycin C (Wood, 2010). Cells defective in other NER genes are comparatively less sensitive to ICL-inducing agents (Andersson et al., 1996; Damia et al., 1996). This shows that ERCC1–XPF has an important role in ICL repair, independent of the NER pathway (Ahmad et al., 2008; Wood, 2010; Fisher et al., 2008; Rahn et al., 2010; Kuraoka et al., 2000). When a DNA replication fork encounters an ICL, a DSB can be formed by enzymatic action (Ahmad et al., 2008; Niedernhofer et al., 2001; Bergstralh and Sekelsky, 2008). XPF and ERCC1 mutants are reported to show defects in the incision or “unhooking” step of ICL repair (De Silva et al., 2000), as well as in the downstream processing of ICL-associated DSBs (De Silva et al., 2002). ERCC1–XPF can cleave 5′ to an ICL in duplex DNA near the junction between single-stranded and double stranded DNA. Following this, it is capable of making a further cut on the other side of the ICL to generate a 4-mer crosslinked to DNA that may be an intermediate for the further processing (Kuraoka et al., 2000; Fisher et al., 2008).
ERCC1–XPF interacts with the SLX4 protein, during the processing of an ICL in cooperation with ubiquitinated FANCD2 (Klein Douwel et al., 2014). SLX4 acts as a scaffold protein during ICL repair. ERCC1–XPF binds to the N-terminal end of SLX4, enhancing action of the nuclease complex in mouse cells and in extracts from Xenopus eggs (Klein Douwel et al., 2014; Hodskinson et al., 2014). The mismatch recognition protein complex MSH2–MSH3 also binds to SLX4 (Svendsen et al., 2009; Kim et al., 2013), and there appear to be direct interactions between ERCC1 and MSH2–MSH3 (Lan et al., 2004).
There appear to be several different routes for ICL repair in mammalian cells; for example, some ICL repair can occur in the absence of DNA replication, as in non-dividing cells (Vasquez, 2010). In bacteria, a psoralen ICL (Van Houten et al., 1986) and a psoralen ICL directed by a triplex-forming oligonucleotide (Christensen et al., 2008) can be repaired by dual incision via the NER pathway. It is possible that this also occurs in mammalian cells. Finally, repair of the unhooked ICL may be completed by either homologous recombination using a new template or by TLS-mediated bypass and NER to remove an unhooked structure (Wood, 2010; Kuraoka et al., 2000; Deans and West, 2011). During ICL repair, ERCC1 may be regulated by USP45-dependent deubiquitination. ERCC1 binds to the USP45 deubiquitylase, and ERCC1 ubiquitination was increased in USP45 knockout cells (Perez-Oliva et al., 2015).
3.4. Base excision repair
Exposure of cells to reactive oxygen and nitrogen species damages the bases and the sugar-phosphate backbone of DNA. Some DNA glycosylases such as OGG1, NEIL1, and NTH1 can remove specific oxidized bases, and then cleave the resulting apurinic/apyrimidinic (AP) site with an associated AP lyase activity. This leaves the 3′ end with a non-hydroxyl group such as a deoxyribose phosphate. The 3′ groups must be removed to allow complete repair. An AP nuclease (in mammalian cells, APEX1) normally acts to convert these 3′ ends to a 3′-OH terminus. In S. cerevisiae, processing of such 3′ ends can still occur in the absence of AP nuclease. Such backup processing is dependent on Rad1–Rad10 (Guillet and Boiteux, 2002). Similarly, mammalian ERCC1–XPF also can remove a 3′-phosphoglycolate (produced by reactive oxygen species attack) from the 3′ end of DNA in vitro (Fisher et al., 2011). ERCC1–XPF may therefore serve a backup role in processing of DNA damaged by reactive oxygen species in mammalian cells.
3.5. Maintenance of telomeres
ERCC1–XPF shows genetic interactions with the telomere maintenance protein TRF2. Mice overexpressing TRF2 are hypersensitive to UV radiation compared to normal mice. Chromosomes from these mice suffer telomere loss that is dependent on ERCC1–XPF (Munoz et al., 2005). On the other hand, ERCC1–XPF expression in TRF2-deficient cells causes increased chromosomal end fusions (Zhu et al., 2003). Mechanistically, ERCC1–XPF can cleave 3′ overhangs from uncapped telomeres. This results in the shortening of the telomeres, together with premature aging in mice. ERCC1–XPF and TRF2 are associated via interaction with the SLX4 protein, and together they participate in regulation of telomere length (Munoz et al., 2005; Zhu et al., 2003; Wu et al., 2008; Wan et al., 2013). The balance between the expression of ERCC1–XPF and TRF2 may be critical for life expectancy and genomic stability.
4. Structural biochemical mechanism and interactions
The human ERCC1 protein has 297 amino acids and a molecular mass of 32,500 Da, while the ERCC4-encoded XPF protein comprises 916 amino acids, with a molecular mass of 104,000 Da. ERCC1–XPF functions as a complex to form an active endonuclease in DNA damage repair. The active nuclease domain residues are present in XPF, but XPF alone is not a nuclease. ERCC1 is necessary as a partner, serving as a critical DNA binding subunit. As a nuclease ERCC1–XPF does not cut double-stranded or single-stranded DNA. Instead, it cuts DNA specifically at junctions between single-stranded and double-stranded DNA, where the single strand departs 5′ to 3′ from the junction. Structures that are cleaved in this manner (with varying efficiencies) include stem-loop bubble substrates, substrates with splayed arms, flaps and protruding single arms (Fig. 1). These represent intermediate structures during many DNA transactions (O'Donovan et al., 1994; Bessho et al., 1997b; de Laat et al., 1998a; Evans et al., 1997b). ERCC1–XPF is the prototype for a family of structure specific-nucleases, which are related by structural and primary sequence similarity. In mammalian cells, members of this family include MUS81-EME1 and SLX4–SLX1, each of which has different and defined DNA substrate preferences (Hanada et al., 2006; Heyer, 2004).
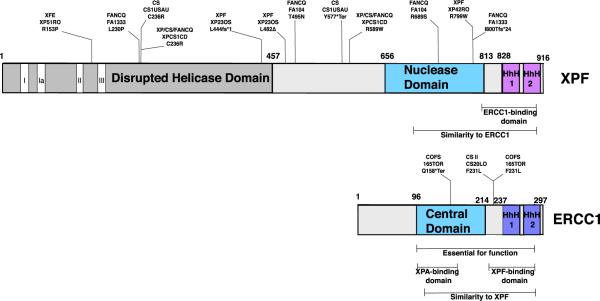
ERCC1 contains a central domain interacting with DNA and proteins, a DNA-interacting helix–hairpin–helix (HhH) domain in the C-terminus, and a region at the N-terminus that is dispensable for some functions (Fig. 2). XPF consists of an N-terminal domain with distant homology to DNA helicases of superfamily 2, a central domain containing the nuclease active site, and a C-terminal HhH domain. The helicase similarity domain of XPF lacks catalytic residues that would confer ATP hydrolysis or nuclease activity, and it can be considered a disrupted helicase retaining only structural homology (Sgouros et al., 1999). ERCC1 and XPF have sequence similarity to one another in their central and nuclease domains, and in their HhH domains (Gaillard and Wood, 2001). It appears that the mammalian ERCC1 gene arose in an ancient gene duplication event involving an ancestor of the ERCC4 gene.
Fig. 2.
Domain arrangement of the ERCC1 and ERCC4/XPF proteins, with examples of mutations found in human syndromes. XPF includes an N-terminal disrupted helicase domain and a nuclease domain. The central domain of ERCC1 shows similarity to the XPF nuclease domain, with critical nuclease residues absent. Both proteins have a C-terminal domain with two helix–hairpin–helix (HhH) motifs.
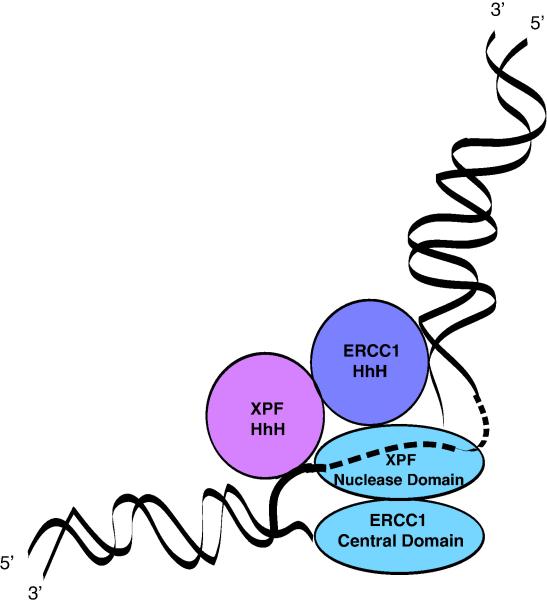
The nuclease domain of XPF includes residues that coordinate a divalent cationic metal atom, and these residues are necessary for nuclease activity (Enzlin and Scharer, 2002). The corresponding homologous region of ERCC1 lacks these conserved metal-coordinating residues (Tsodikov et al., 2005). ERCC1 and XPF interact via their similar C-terminal helix–hairpin–helix domains (de Laat et al., 1998a,b; Tsodikov et al., 2005). In addition to dimerization, the HhH domains also contribute to DNA binding (de Laat et al., 1998c; Doherty et al., 1996; Su et al., 2012). The central domain of ERCC1 binds to single-stranded DNA with higher affinity than to double-stranded DNA (Tsodikov et al., 2005; McNeil and Melton, 2012). It has been proposed that the central domain of ERCC1 binds to single-stranded DNA, while the HhH regions of ERCC1 and XPF secure two splayed arms of DNA to test and insure a conformation allowing nuclease activity (Tsodikov et al., 2005) (Fig. 3). Structural studies by NMR indicate that the HhH2 domain of XPF binds to ssDNA, and the HhH2 domain of ERCC1 binds to dsDNA (Das et al., 2012).
Fig. 3.
ERCC1 and XPF interaction and binding of the complex with DNA (Friedberg et al., 2006). The central domain of ERCC1 binds to single-stranded DNA regions. The HhH regions of ERCC1 and XPF associate with one another, and bind to DNA to position the nuclease domain of XPF near the junction between single-stranded and double-stranded DNA.
A minimal complex consisting of N-terminally truncated ERCC1 beginning at residue 95 and N-terminally truncated XPF beginning at residue 656 can mediate some structure-specific cleavage of DNA (Tsodikov et al., 2005), showing that the N-terminal helicase-like domain of XPF is not absolutely required for the activity. However, the Arg153Pro substitution found in the XFE progeroid patient described below causes hypersensitivity to UV and ICL causing agents (Niedernhofer et al., 2006). This underlines that the conformation of the N-terminal region of XPF influences ERCC1–XPF activity.
Interactions with different protein partners are specific for of the pathway where ERCC1–XPF is involved. During NER, ERCC1 interacts with XPA (Li et al., 1994; Park and Sancar, 1994), while the N-terminal region of XPF binds to RPA. Further, RPA directly interacts with XPA. These interactions are necessary to target the ERCC1–XPF nuclease for specific action during NER (Li et al., 1994; Bessho et al., 1997b; McNeil and Melton, 2012; Matsunaga et al., 1996; Orelli et al., 2010). During ICL repair, ERCC1–XPF interacts with SLX4 (Klein Douwel et al., 2014; Hodskinson et al., 2014).
Formation of the ERCC1–XPF complex is also necessary to maintain stability of the two proteins (Gaillard and Wood, 2001; Sijbers et al., 1996b). When the individual subunits of ERCC1 and XPF are expressed separately, they self-aggregate in the absence of the other partner (Gaillard and Wood, 2001), a condition that would lead to proteolytic degradation in mammalian cells. Extracts of yeast S. cerevisiae Rad1 or Rad10 mutant cells can be complemented to nearly normal levels by introduction of the missing protein (Wang et al., 1993). However, this is not the case for human ERCC1 and XPF mutants (Gaillard and Wood, 2001). Effectively, XPF mutant cells are also ERCC1-deficient, and ERCC1 mutant cells are XPF-deficient (Gaillard and Wood, 2001; Bhagwat et al., 2009; Niedernhofer et al., 2007).
5. Disorders associated with ERCC1-XPF
Several human inherited disorders are associated with mutations in the ERCC1 or ERCC4 genes. As described below, these include xeroderma pigmentosum, Cockayne syndrome, Fanconi anemia, XFE progeria, and cerebro-oculo-facio-skeletal syndrome. Mouse models for ERCC1 and XPF deficiency are proving valuable in understanding these conditions.
5.1. Disorders associated with XPF
5.1.1. Xeroderma pigmentosum
A rare autosomal disorder, xeroderma pigmentosum (XP) is characterized by extreme sensitivity to sunlight, pigmentation abnormalities, and predisposition to skin cancer due to deficiencies in the NER pathway (Friedberg et al., 2006; Kraemer and DiGiovanna, 1993; Cleaver, 2005; Lehmann et al., 2011; DiGiovanna and Kraemer, 2012). Abnormalities include photosensitivity, cutaneous atrophy, cutaneous telangiectasia, actinic keratoses, and malignant skin neoplasms (Friedberg et al., 2006). Neurological abnormalities, observed in a minority of cases, include low intelligence, abnormal motor activity, areflexia, impaired hearing, abnormal speech, and microcephaly. Ocular abnormalities include conjunctival injection, corneal abnormalities, impaired vision, photophobia, and neoplasms. Eight complementation groups (XP-A to XP-G, and XP-V) have been classified.
XP-F, occurring less frequently overall than other XP cases, accounts for approximately 7% of reported cases in Japan (Kraemer and DiGiovanna, 1993). Severe ocular and neurological abnormalities are rare in the XP-F patients observed (Gregg et al., 2011). Missense mutations are present in at least one allele in all XP-F patients, none of which affect the nuclease activity of the enzyme (Gregg et al., 2011). Basal cell carcinomas, squamous cell carcinomas, and keratoacanthomas were found in several diagnosed XP-F patients (Sijbers et al., 1998a).
One case, not typical of other XP-F cases, is patient XP42RO. This patient had mild ocular photophobia and exhibited acute skin reactions upon exposure to sunlight (Sijbers et al., 1998a). Basal and squamous cell carcinomas appeared after twenty-seven years of age. Progressive neurologic symptoms emerged in his late forties, which is unusual for an XP-F patient. XP42RO was homozygous for a R799W point mutation, a mutation that has also been identified in at least eight other XP-F patients (de Laat et al., 1998c; Gregg et al., 2011) (Fig. 2).
5.1.2. XFE progeroid syndrome
A progeroid syndrome has been described resulting from a mutation in the ERCC4 gene (Niedernhofer et al., 2006). The patient, XP51RO, exhibited symptoms resembling premature aging, profound crosslink sensitivity, and frequent sunburns. Many neurologic, hepatobiliary, musculoskeletal, and hematopoietic symptoms were present. This clinical presentation is distinct from xeroderma pigmentosum, Cockayne syndrome, or combined xeroderma pigmentosum–Cockayne syndrome. XP51RO cells harbor a homozygous C → G transversion affecting position 458 in ERCC4, yielding a R153P mutation at a conserved arginine (Table 2). The mutation is located between motifs II and III of the disrupted helicase domain in a leucine rich region which may be involved in protein–protein interactions (Tsodikov et al., 2005; Niedernhofer et al., 2006) (Fig. 2).
Table 2.
Examples of ERCC1 and ERCC4 mutations in human individuals with various inherited disorders.
| Disorder | Patient | Ref. | Protein | Mutation | Protein change |
|---|---|---|---|---|---|
| FA | FA104 | Bogliolo et al. (2013) | XPF | 5 bp deletion exon 8 | Thr495Asnfs*6 |
| Missense mutation exon 11 | Arg689Ser | ||||
| FA | 1333 | Bogliolo et al. (2013) | XPF | 28 bp duplication in exon 11 of maternal allele | Ile800Thrfs*24 |
| Missense mutation in paternal allele | Leu230Pro | ||||
| CS | CS1USAU | Kashiyama et al. (2013) | XPF | Exon 4 point mutation | Cys236Arg |
| Exon 8 frameshift insertion, premature stop codon | Tyr577* | ||||
| XP/CS/FA | XPCS1CD | Kashiyama et al. (2013) | XPF | Heterozygous point mutation | Cys236Arg |
| Heterozygous point mutation | Arg589Trp | ||||
| XFE progeroid | XP51RO | Niedernhofer et al. (2006) | XPF | Point mutation | Arg153Pro |
| XP | XP23OS | Matsumura et al. (1998) | XPF | 1 bp insertion yielding a frameshift at 444 and stop at 482 | Lys444 and 482* |
| XP | XP42RO | Sijbers et al. (1998b) | XPF | Point mutation | Arg799Trp |
| COFS | 165TOR | Jaspers et al. (2007) | ERCC1 | Point mutation (Maternal allele) | Gln158* |
| Point mutation (Paternal allele) | Phe231Leu | ||||
| CS type II | CS20LO(103) | Kashiyama et al. (2013) | ERCC1 | Homozygous mutation, exon 7 | Phe231Leu |
indicates that the mutation produces a termination codon, sometimes with a frameshift (fs) resulting in the indicated number of amino acid residues before termination.
5.1.3. Fanconi anemia and Cockayne syndrome
Fanconi anemia (FA) is distinguished by bone marrow failure, predisposition to cancer, congenital malformations, hypersensitivity to crosslinking agents, and chromosome fragility. Seventeen genes have been identified as FA genes. Most recently ERCC4 was identified as a FA gene, designated FANCQ in that context (Bogliolo et al., 2013). Patient FA104 was diagnosed with a malformative syndrome indicating FA, and developed bone marrow failure when 2 years old. The ERCC4 gene of patient FA104 contained a 5 bp deletion in exon 8 (c.1484_1488 delCTCAA), leading to a frameshift and premature stop codon, as well as a missense mutation in exon 11 leading to an Arg689Ser change (Fig. 2, Table 2). Lymphoblastoid cell lines from this patient were sensitive to mitomycin-C and melphalan, which is consistent with FA, but were not hypersensitive to topoisomerase I inhibitor camptothecin or PARP inhibitor KU58948. FA104 cells showed normal FANCD2 monoubiquitination and RAD51 foci formation (Bogliolo et al., 2013).
Patient FA1333 was diagnosed with FA at 5 years of age. Symptoms included various developmental defects and bone marrow failure (Bogliolo et al., 2013). A 28 bp duplication is present in exon 11 of the maternal allele and is predicted to encode for a truncated XPF that lacks the double helix–hairpin–helix domain involved in ERCC1 and DNA binding interactions (Fig. 2). A missense mutation resulting in Leu to Pro at residue 230 is present in the paternal allele, causing a mutation in the disrupted helicase domain (Fig. 2). FA1333 lymphoblastoid cell lines derived from this patient were not impaired in FANCD2 monoubiquitination or RAD51 foci formation. Cells were sensitive to mitomycin C and melphalan, but insensitive to camptothecin and KU58948 (Bogliolo et al., 2013).
Cockayne syndrome (CS) patients suffer from developmental defects, neurological abnormalities, and abnormal skin photosensitivity (Kashiyama et al., 2013). CS can be classified into three different groups, CS type I, CS type II, and CS type III, based on severity. Sunlight sensitivity is milder than that of XP cases and skin cancers are not found in CS. Most CS individuals have mutations in ERCC8 (CSA) or ERCC6 (CSB), resulting in defective TC-NER. ERCC1 or ERCC4 mutations have been reported in at least three CS cases where ERCC6 and ERCC8 function was normal (Kashiyama et al., 2013).
Patient XPCS1CD displayed many characteristics of CS, XP, and FA, with exceptional sensitivity to mitomycin C (Lehmann et al., 2011). Symptoms include various developmental defects, neurological defects, sunburns with minimal sun exposure, and abnormal bone marrow (Lehmann et al., 2011). XP/CS/FA patient XPCS1CD carried two ERCC4 heterozygous point mutations, yielding Cys236Arg and Arg589Trp variants (Fig. 2).
Cockayne syndrome patient CS1USAU had two heterozygous mutations in ERCC4: a point mutation yielding Cys236Arg (also found in XPCS1CD) located in the SF2 disrupted helicase domain and an insertion resulting in premature stop codon Tyr577* (Kashiyama et al., 2013) (Fig. 2). Dermal fibroblasts isolated from the patient showed a reduction in RNA-synthesis activity, signifying a deficiency in TC-NER (Lehmann et al., 2011).
5.1.4. ERCC4 mouse models
In the cDNA of human patient XP23OS, an adenine insertion was detected leading to a frameshift mutation and ensuing stop codon that would lead to a deletion of the C-terminal half of XPF (Table 2) (Tian et al., 2004; Matsumura et al., 1998). It was hypothesized that XP23OS may be homozygous for this mutation or that the other allele is transcriptionally repressed. The only marked symptom exhibited by this patient was photosensitivity of the skin. XPF mutant mice mimicking the XP23OS patient mutation were generated by engineering a stop codon in exon 8, resulting in truncation of XPF at residue 445 (Tian et al., 2004). Approximately 15 days after birth, the weight of homozygous mice was 27% of wild-type controls or heterozygous mutants. Homozygous Ercc4 mutant mice died at approximately 3 weeks. Their organs appeared morphologically normal, yet significantly smaller (Tian et al., 2004). However, hepatocytes from homozygous mutant mice contained enlarged nuclei. This severe phenotype is similar to that of mice with inactivating mutations in ERCC1, as described below. It is possible that the relatively mild phenotype of the human XP23OS patient arose because the second XPF allele in this individual expressed a small amount of normal XPF protein, or XPF protein with a less debilitating mutation.
5.2. Disorders associated with ERCC1 mutations
5.2.1. Cerebro-oculo-facio-skeletal syndrome/Cockayne syndrome
Individuals diagnosed with cerebro-oculo-facio-skeletal syndrome (COFS) have been identified with defects in NER genes due to mutations in ERCC6/CSB, ERCC2/XPD, or ERCC5/XPG (Meira et al., 2000; Graham et al., 2001; Hamel et al., 1996). In addition, the first patient (165TOR) diagnosed with ERCC1 deficiency suffered from COFS. This individual exhibited severe developmental failure, mild NER impairment, microcephaly, and other facial and skeletal abnormalities (Jaspers et al., 2007). ERCC1 cDNA and genomic DNA from 165TOR cells harbors a C → T transition, yielding a premature Q158X termination mutation derived from the maternal allele (Fig. 2). AC → G transversion in the paternal allele results in a Phe to Leu mutation at residue 231, which lies within the XPF-interacting helix–hairpin–helix domain (Kashiyama et al., 2013). This residue is normally conserved as Phe or Tyr in mammals, fish, plants, and fungi.
Cockayne syndrome type II patient CS20LO exhibited a homozygous mutation in exon 7 of ERCC1, producing a F231L mutation, which was also observed in patient 165TOR (Fig. 2).
5.2.2. ERCC1 mouse models
Several mouse models have been generated to study the consequences of disrupting Ercc1 function (Gregg et al., 2011). In one model (Ercc1tm1Dwm), the last four exons are lacking due to a mutation in exon 5, producing a truncated ERCC1 protein that is unable to bind XPF (Gregg et al., 2011; McWhir et al., 1993). This Ercc1 mutation resulted in elevated p53 levels in liver, brain, and kidney in the perinatal period. Mice died before weaning and had liver nuclear abnormalities (McWhir et al., 1993). In another model (Ercc1tmJhjh), a neomycin resistance cassette was inserted into exon 7, resulting in the truncation of the helix–hairpin–helix motif required for XPF interaction (Gregg et al., 2011; Weeda et al., 1997). Postnatal growth was severely retarded and the mice died at approximately 3 weeks. Hematopoietic cells undergo rapid turnover, leading to premature exhaustion of stem cell reserves in Ercc1-deficient mice. Mice lacking ERCC1 also had musculoskeletal and nervous system defects, and impaired liver function (Niedernhofer et al., 2006; Gregg et al., 2011).
Mutational analysis has shown that NER and crosslink repair are mildly affected by the deletion of the last four amino acids of ERCC1, and that repair is severely impaired by the truncation of the last five or six residues (Sijbers et al., 1996b). Residues 293 to 297 are at the C-terminal border of the HhH XPF binding domain (de Laat et al., 1998c). Removal of the first 91 amino acids does not significantly impact ERCC1 function (Sijbers et al., 1996b). An Ercc1 mutant mouse was generated containing a seven amino-acid carboxy-terminal truncation (Ercc1292 or Ercc1Δ) (Weeda et al., 1997). Ercc1Δ homozygous mutants were born at lower than expected Mendelian ratio, were severely runted, and had a reduced lifespan. Ercc1Δ mice in the FVB mouse strain background had a maximal age of 78 days (Weeda et al., 1997).
ERCC1-deficient mice exhibit many signs of accelerated aging. Ercc1−/Δ mice express 10% of the ERCC1–XPF content found in wild-type mice (Goss et al., 2011). These mice have a maximum lifespan of 32 weeks, while exhibiting marked premature age-related changes throughout the animal. Osteoporosis and intervertebral disc degeneration occur in Ercc1−/Δ mice, with disc height reduced by 20–30% in 20-week old Ercc1−/Δ mice in comparison to wild type controls (Gregg et al., 2011; Vo et al., 2010). Disc height of Ercc1−/Δ mice at 20 weeks was comparable to naturally aging 2-year old mice. Ercc1−/Δ mice displayed an age-dependent loss of disc matrix proteoglycans, which are critical for counteracting spine compression. Loss of proteoglycans in these mice may be partially attributed to increased cell senescence and apoptosis, which are responses to DNA damage (Vo et al., 2010). Ercc1−/Δ mice are also afflicted with peripheral neuropathy, a degenerative disorder often associated with aging (Goss et al., 2011). Nerve conduction studies revealed significant abnormalities in 20-week old Ercc1−/Δ mice, and the mice exhibit loss of peripheral nerve fibers, abnormal myelin structures, and myelin degeneration. This indicates that these mice both spontaneously and prematurely acquire peripheral sensory and motor neuropathy. Due to the fact that Ercc1−/Δ mice are deficient in multiple DNA repair pathways, these data support the concept that DNA damage may contribute to age related neurodegeneration (Goss et al., 2011). Gene expression analysis of XPF–ERCC1 deficient mice indicates increased cell death and antioxidant defenses, a shift towards anabolism and reduced growth hormone/insulin-like growth factor 1 signaling (Niedernhofer et al., 2006). Similar changes are seen in wild-type mice in response to chronic genotoxic stress, caloric restriction, or with aging.
6. ERCC1–XPF and cancer treatment
As expected, reduction of ERCC1–XPF protein by shRNA-mediated suppression sensitizes cells to killing by DNA crosslinking agents such as cisplatin derivatives (Arora et al., 2010), which are extensively used in treatment of breast, prostate, ovarian and other cancers. Thus a combination of ERCC1–XPF inhibition together with chemotherapy may be effective for the treatment of some cancers. For this reason, it has been important to identify antibodies that can be used for immunohistochemical identification of ERCC1 and XPF in formalin-fixed, paraffin embedded sections (Bhagwat et al., 2009; Smith et al., 2014). Several approaches have been proposed to disrupt the function of ERCC1–XPF complex (reviewed in (McNeil and Melton, 2012)). These include disruption of the dimerization of ERCC1 and XPF inhibition of XPF nuclease activity, and inhibition of the binding of ERCC1–XPF to DNA. Disruption of protein–protein interactions is another possibility, targeting interactions between ERCC1 and XPA or between XPF and RPA, SLX4, or RAD52 (reviewed in (McNeil and Melton, 2012)). Cells defective in ERCC1–XPF are more sensitive than normal to inhibitors of the ATR protein (Mohni et al., 2014), and to inhibitors of poly(ADP-ribose) polymerase (Postel-Vinay et al., 2013). These observations might open additional possibilities for combination therapies.
It is possible that high expression of ERCC1–XPF could be associated with resistance to some DNA damaging drugs. Few studies have been performed with antibodies verified to recognize ERCC1–XPF specifically and uniquely, but better reagents are now available that could be used to quantify the expression of ERCC1–XPF in patients before the treatment with such drugs (Bhagwat et al., 2009; Smith et al., 2014). Levels of both the ERCC1 and the XPF proteins would have to be elevated in order to have any biological effect.
Acknowledgments
This review and the corresponding Gene Wiki article are written as part of the Gene Review a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Wiki Initiative is supported by National Institute of Health (GM089820). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. We thank our research group colleagues for comments on the manuscript. Research in RDW's laboratory was supported by National Institutes of Health (NIH) grants CA132840, CA09717 from the National Cancer Institute, the M.D. Anderson Research Trust and the Grady F. Saunders Ph.D. Distinguished Research Professorship. We also acknowledge funding from NIH Cancer Center Support Grant P30-CA016672 to the University of Texas MD Anderson Cancer Center.
Abbreviations
- AP
apurinic/apyrimidinic
- ATP
adenosine triphosphate
- cDNA
complementary deoxyribonucleic acid
- CHO
Chinese hamster ovary
- COFS
cerebro-oculo-facio-skeletal syndrome
- CS
Cockayne syndrome
- DSB
double strand break
- FA
Fanconi anemia
- GG-NER
global genome nucleotide excision repair
- HhH
helix–hairpin–helix
- ICL
interstrand crosslink in DNA
- kb
kilobase
- NER
nucleotide excision repair
- NHEJ
non-homologous end joining
- OB-fold
oligonucleotide/oligosaccharide binding fold, the basic structural unit of binding to single-stranded DNA
- RPA
replication protein A
- SF2
helicases of superfamily 2
- shRNA
small hairpin ribonucleic acid
- TC-NER
transcription coupled nucleotide excision repair
- TFIIH
transcription factor II H
- UV
ultraviolet
- XP
xeroderma pigmentosum
- γH2AX
H2A histone family, member X, phosphorylated at serine 139
References
- Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- Adair GM, Rolig RL, Moore-Faver D, Zabelshansky M, Wilson JH, Nairn RS. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 2000;19:5552–5561. doi: 10.1093/emboj/19.20.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH, Niedernhofer LJ. ERCC1–XPF endonuclease facilitates DNA double-strand break repair. Mol. Cell. Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36:1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Minawi AZ, Lee YF, Hakansson D, Johansson F, Lundin C, Saleh-Gohari N, Schultz N, Jenssen D, Bryant HE, Meuth M, et al. The ERCC1/XPF endonuclease is required for completion of homologous recombination at DNA replication forks stalled by inter-strand cross-links. Nucleic Acids Res. 2009;37:6400–6413. doi: 10.1093/nar/gkp705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson BS, Sadeghi T, Siciliano MJ, Legerski R, Murray D. Nucleotide excision repair genes as determinants of cellular sensitivity to cyclophosphamide analogs. Cancer Chemother. Pharmacol. 1996;38:406–416. doi: 10.1007/s002800050504. [DOI] [PubMed] [Google Scholar]
- Arora S, Kothandapani A, Tillison K, Kalman-Maltese V, Patrick SM. Downregulation of XPF–ERCC1 enhances cisplatin efficacy in cancer cells. DNA Repair (Amst) 2010;9:745–753. doi: 10.1016/j.dnarep.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell AJ, Bardwell L, Johnson DK, Friedberg EC. Yeast DNA recombination and repair proteins Rad1 and Rad10 constitute a complex in vivo mediated by localized hydrophobic domains. Mol. Microbiol. 1993;8:1177–1188. doi: 10.1111/j.1365-2958.1993.tb01662.x. [DOI] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstralh DT, Sekelsky J. Interstrand crosslink repair: can XPF–ERCC1 be let off the hook? Trends Genet. 2008;24:70–76. doi: 10.1016/j.tig.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Bessho T, Mu D, Sancar A. Initiation of DNA interstrand cross-link repair in humans: the nucleotide excision repair system makes dual incisions 5′ to the cross-linked base and removes a 22- to 28-nucleotide-long damage-free strand. Mol. Cell. Biol. 1997a;17:6822–6830. doi: 10.1128/mcb.17.12.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho T, Sancar A, Thompson LH, Thelen MP. Reconstitution of human excision nuclease with recombinant XPF–ERCC1 complex. J. Biol. Chem. 1997b;272:3833–3837. doi: 10.1074/jbc.272.6.3833. [DOI] [PubMed] [Google Scholar]
- Bhagwat NR, Roginskaya VY, Acquafondata MB, Dhir R, Wood RD, Niedernhofer LJ. Immunodetection of DNA repair endonuclease ERCC1–XPF in human tissue. Cancer Res. 2009;69:6831–6838. doi: 10.1158/0008-5472.CAN-09-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggerstaff M, Szymkowski DE, Wood RD. Co-correction of the ERCC1, ERCC4 and xeroderma pigmentosum group F DNA repair defects in vitro. EMBO J. 1993;12:3685–3692. doi: 10.1002/j.1460-2075.1993.tb06043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, Raams A, Trujillo JP, Minguillon J, Ramirez MJ, Pujol R, et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am. J. Hum. Genet. 2013;92:800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookman KW, Lamerdin JE, Thelen MP, Hwang M, Reardon JT, Sancar A, Zhou ZQ, Walter CA, Parris CN, Thompson LH. ERCC4 (XPF) encodes a human nucleotide excision repair protein with eukaryotic recombination homologs. Mol. Cell. Biol. 1996;16:6553–6562. doi: 10.1128/mcb.16.11.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen LA, Wang H, Van Houten B, Vasquez KM. Efficient processing of TFO-directed psoralen DNA interstrand crosslinks by the UvrABC nuclease. Nucleic Acids Res. 2008;36:7136–7145. doi: 10.1093/nar/gkn880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer. 2005;5:564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- Damia G, Imperatori L, Stefanini M, D'Incalci M. Sensitivity of CHO mutant cell lines with specific defects in nucleotide excision repair to different anti-cancer agents. Int. J. Cancer. 1996;66:779–783. doi: 10.1002/(SICI)1097-0215(19960611)66:6<779::AID-IJC12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Das D, Folkers GE, van Dijk M, Jaspers NG, Hoeijmakers JH, Kaptein R, Boelens R. The structure of the XPF-ssDNA complex underscores the distinct roles of the XPF and ERCC1 helix–hairpin–helix domains in ss/ds DNA recognition. Structure. 2012;20:667–675. doi: 10.1016/j.str.2012.02.009. [DOI] [PubMed] [Google Scholar]
- de Laat WL, Appeldoorn E, Jaspers NG, Hoeijmakers JH. DNA structural elements required for ERCC1–XPF endonuclease activity. J. Biol. Chem. 1998a;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- de Laat WL, Appeldoorn E, Sugasawa K, Weterings E, Jaspers NGJ, Hoeijmakers JHJ. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 1998b;12:2598–2609. doi: 10.1101/gad.12.16.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat WL, Sijbers AM, Odijk H, Jaspers NGJ, Hoeijmakers JHJ. Mapping of interaction domains between human repair proteins ERCC1 and XPF. Nucleic Acids Res. 1998c;26:4146–4152. doi: 10.1093/nar/26.18.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defects in interstrand cross-link uncoupling do not account for the extreme sensitivity of ERCC1 and XPF cells to cisplatin. Nucleic Acids Res. 2002;30:3848–3856. doi: 10.1093/nar/gkf479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J. Invest. Dermatol. 2012;132:785–796. doi: 10.1038/jid.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty AJ, Serpell LC, Ponting CP. The helix–hairpin–helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enzlin JH, Scharer OD. The active site of the DNA repair endonuclease XPF– ERCC1 forms a highly conserved nuclease motif. EMBO J. 2002;21:2045–2053. doi: 10.1093/emboj/21.8.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Moggs JG, Hwang JR, Egly J-M, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997a;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Fellows J, Coffer A, Wood RD. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 1997b;16:625–638. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher LA, Bessho M, Bessho T. Processing of a psoralen DNA interstrand cross-link by XPF–ERCC1 complex in vitro. J. Biol. Chem. 2008;283:1275–1281. doi: 10.1074/jbc.M708072200. [DOI] [PubMed] [Google Scholar]
- Fisher LA, Samson L, Bessho T. Removal of reactive oxygen species-induced 3′-blocked ends by XPF–ERCC1. Chem. Res. Toxicol. 2011;24:1876–1881. doi: 10.1021/tx200221j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell J, Haber JE. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2nd ed. ASM Press; Washington, DC.: 2006. [Google Scholar]
- Gaillard PH, Wood RD. Activity of individual ERCC1 and XPF subunits in DNA nucleotide excision repair. Nucleic Acids Res. 2001;29:872–879. doi: 10.1093/nar/29.4.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss JR, Stolz DB, Robinson AR, Zhang M, Arbujas N, Robbins PD, Glorioso JC, Niedernhofer LJ. Premature aging-related peripheral neuropathy in a mouse model of progeria. Mech. Ageing Dev. 2011;132:437–442. doi: 10.1016/j.mad.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JM, Jr., Anyane-Yeboa K, Raams A, Appeldoorn E, Kleijer WJ, Garritsen VH, Busch D, Edersheim TG, Jaspers NG. Cerebro-oculo-facio-skeletal syndrome with a nucleotide excision-repair defect and a mutated XPD gene, with prenatal diagnosis in a triplet pregnancy. Am. J. Hum. Genet. 2001;69:291–300. doi: 10.1086/321295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg SQ, Robinson AR, Niedernhofer LJ. Physiological consequences of defects in ERCC1–XPF DNA repair endonuclease. DNA Repair (Amst) 2011;10:781–791. doi: 10.1016/j.dnarep.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet M, Boiteux S. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J. 2002;21:2833–2841. doi: 10.1093/emboj/21.11.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel BC, Raams A, Schuitema-Dijkstra AR, Simons P, van der Burgt I, Jaspers NG, Kleijer WJ. Xeroderma pigmentosum–Cockayne syndrome complex: a further case. J. Med. Genet. 1996;33:607–610. doi: 10.1136/jmg.33.7.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of inter-strand DNA crosslinks into double-strands breaks. EMBO J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Heyer WD. Recombination: Holliday junction resolution and crossover formation. Curr. Biol. 2004;14:R56–R58. doi: 10.1016/j.cub.2003.12.043. [DOI] [PubMed] [Google Scholar]
- Hodskinson MR, Silhan J, Crossan GP, Garaycoechea JI, Mukherjee S, Johnson CM, Scharer OD, Patel KJ. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF–ERCC1 in DNA crosslink repair. Mol. Cell. 2014;54:472–484. doi: 10.1016/j.molcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JC, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl. Acad. Sci. U. S. A. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Haber JE. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:2245–2251. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers NG, Raams A, Silengo MC, Wijgers N, Niedernhofer LJ, Robinson AR, Giglia-Mari G, Hoogstraten D, Kleijer WJ, Hoeijmakers JH, et al. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am. J. Hum. Genet. 2007;80:457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiyama K, Nakazawa Y, Pilz DT, Guo C, Shimada M, Sasaki K, Fawcett H, Wing JF, Lewin SO, Carr L, et al. Malfunction of nuclease ERCC1–XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am. J. Hum. Genet. 2013;92:807–819. doi: 10.1016/j.ajhg.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Spitz GS, Veturi U, Lach FP, Auerbach AD, Smogorzewska A. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood. 2013;121:54–63. doi: 10.1182/blood-2012-07-441212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Raschle M, Walter JC, Knipscheer P. XPF–ERCC1 acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol. Cell. 2014;54:460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer KH, DiGiovanna JJ. In: Pagon RA, Adam MP, Ardinger HH, Bird TD, Dolan CR, Fong CT, Smith RJH, Stephens K, editors. GeneReviews(R), Seattle (WA): 1993. [Google Scholar]
- Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA crosslink initiated by ERCC1–XPF repair/recombination nuclease. J. Biol. Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- Lan L, Hayashi T, Rabeya RM, Nakajima S, Kanno S, Takao M, Matsunaga T, Yoshino M, Ichikawa M, Riele H, et al. Functional and physical interactions between ERCC1 and MSH2 complexes for resistance to cis-diamminedichloroplatinum(II) in mammalian cells. DNA Repair (Amst) 2004;3:135–143. doi: 10.1016/j.dnarep.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Bootsma D, Clarkson SG, Cleaver JE, McAlpine PJ, Tanaka K, Thompson LH, Wood RD. Nomenclature of human DNA repair genes. Mutat. Res. 1994;315:41–42. doi: 10.1016/0921-8777(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Lehmann AR, McGibbon D, Stefanini M. Xeroderma pigmentosum. Orphanet J. Rare Dis. 2011;6:70. doi: 10.1186/1750-1172-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Elledge SJ, Peterson CA, Bales ES, Legerski RJ. Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl. Acad. Sci. U. S. A. 1994;91:5012–5016. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Siciliano J, White B, Legerski R, Callen D, Reeders S, Siciliano MJ, Thompson LH. Regional mapping of human DNA excision repair gene ERCC4 to chromo-some 16p13.13-p13.2. Mutagenesis. 1993;8:199–205. doi: 10.1093/mutage/8.3.199. [DOI] [PubMed] [Google Scholar]
- Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y, Nishigori C, Yagi T, Imamura S, Takebe H. Characterization of molecular defects in xeroderma pigmentosum group F in relation to its clinically mild symptoms. Hum. Mol. Genet. 1998;7:969–974. doi: 10.1093/hmg/7.6.969. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Park CH, Bessho T, Mu D, Sancar A. Replication protein-A confers structure-specific endonuclease activities to the XPF–ERCC1 and XPG subunits of human DNA-repair excision nucleases. J. Biol. Chem. 1996;271:11047–11050. doi: 10.1074/jbc.271.19.11047. [DOI] [PubMed] [Google Scholar]
- McNeil EM, Melton DW. DNA repair endonuclease ERCC1–XPF as a novel therapeutic target to overcome chemoresistance in cancer therapy. Nucleic Acids Res. 2012;40:9990–10004. doi: 10.1093/nar/gks818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhir J, Selfridge J, Harrison DJ, Squires S, Melton DW. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat. Genet. 1993;5:217–224. doi: 10.1038/ng1193-217. [DOI] [PubMed] [Google Scholar]
- Meira LB, Graham JM, Greenberg CR, Busch DB, Doughty ATB, Ziffer DW, Coleman DM, SavreTrain I, Friedberg EC. Manitoba aboriginal kindred with original cerebro-oculo-facio-skeletal syndrome has a mutation in the Cockayne syndrome group B (CSB) gene. Am. J. Hum. Genet. 2000;66:1221–1228. doi: 10.1086/302867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KD, Ferguson DO, Essers J, Eckersdorff M, Kanaar R, Alt FW. Rad54 and DNA Ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev. 2004;18:1283–1292. doi: 10.1101/gad.1204304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moggs JG, Yarema KJ, Essigmann JM, Wood RD. Analysis of incision sites produced by human cell extracts and purified proteins during nucleotide excision repair of a 1,3-intrastrand d(GpTpG)-cisplatin adduct. J. Biol. Chem. 1996;271:7177–7186. doi: 10.1074/jbc.271.12.7177. [DOI] [PubMed] [Google Scholar]
- Mohni KN, Kavanaugh GM, Cortez D. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer Res. 2014;74:2835–2845. doi: 10.1158/0008-5472.CAN-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu D, Hsu DS, Sancar A. Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem. 1996;271:8285–8294. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- Munoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 2005;37:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- Murray D, Rosenberg E. The importance of the ERCC1/ERCC4[XPF] complex for hypoxic-cell radioresistance does not appear to derive from its participation in the nucleotide excision repair pathway. Mutat. Res. DNA Repair. 1996;364:217–226. doi: 10.1016/s0921-8777(96)00036-5. [DOI] [PubMed] [Google Scholar]
- Murray D, MacAnn A, Hanson J, Rosenberg E. ERCC1/ERCC4 5′-endonuclease activity as a determinant of hypoxic-cell radiosensitivity. Int. J. Radiat. Biol. 1995;69:319–327. doi: 10.1080/095530096145878. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1–Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 2001;20:6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Bhagwat N, Wood RD. ERCC1 and non-small-cell lung cancer. New Engl. J. Med. 2007;356:2538–2540. doi: 10.1056/NEJMc070742. [DOI] [PubMed] [Google Scholar]
- O'Donovan A, Davies AA, Moggs JG, West SC, Wood RD. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- Orelli B, McClendon TB, Tsodikov OV, Ellenberger T, Niedernhofer LJ, Scharer OD. The XPA-binding domain of ERCC1 is required for nucleotide excision repair but not other DNA repair pathways. J. Biol. Chem. 2010;285:3705–3712. doi: 10.1074/jbc.M109.067538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Sancar A. Formation of a ternary complex by human XPA, ERCC1, and ERCC4(XPF) excision-repair proteins. Proc. Natl. Acad. Sci. U. S. A. 1994;91:5017–5021. doi: 10.1073/pnas.91.11.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Oliva AB, Lachaud C, Szyniarowski P, Munoz I, Macartney T, Hickson I, Rouse J, Alessi DR. USP45 deubiquitylase controls ERCC1–XPF endonuclease-mediated DNA damage responses. EMBO J. 2015;34:326–343. doi: 10.15252/embj.201489184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postel-Vinay S, Bajrami I, Friboulet L, Elliott R, Fontebasso Y, Dorvault N, Olaussen KA, Andre F, Soria JC, Lord CJ, et al. A high-throughput screen identifies PARP1/2 inhibitors as a potential therapy for ERCC1-deficient non-small cell lung cancer. Oncogene. 2013;32:5377–5387. doi: 10.1038/onc.2013.311. [DOI] [PubMed] [Google Scholar]
- Prado F, Aguilera A. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn JJ, Adair GM, Nairn RS. Multiple roles of ERCC1–XPF in mammalian inter-strand crosslink repair. Environ. Mol. Mutagen. 2010;51:567–581. doi: 10.1002/em.20583. [DOI] [PubMed] [Google Scholar]
- Rubin JS, Joyner AL, Bernstein A, Whitmore GF. Molecular identification of a human DNA repair gene following DNA-mediated gene transfer. Nature. 1983;306:206–208. doi: 10.1038/306206a0. [DOI] [PubMed] [Google Scholar]
- Sargent RG, Rolig RL, Kilburn AE, Adair GM, Wilson JH, Nairn RS. Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13122–13127. doi: 10.1073/pnas.94.24.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader CE, Vardo J, Linehan E, Twarog MZ, Niedernhofer LJ, Hoeijmakers JH, Stavnezer J. Deletion of the nucleotide excision repair gene Ercc1 reduces immunoglobulin class switching and alters mutations near switch recombination junctions. J. Exp. Med. 2004;200:321–330. doi: 10.1084/jem.20040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgouros J, Gaillard P-HL, Wood RD. A relationship between a DNA-repair/ recombination nuclease family and archaeal helicases. Trends Biochem. Sci. 1999;24:95–97. doi: 10.1016/s0968-0004(99)01355-9. [DOI] [PubMed] [Google Scholar]
- Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei Y-F, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996a;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- Sijbers AM, van der Spek PJ, Odijk H, van den Berg J, van Duin M, Westerveld A, Jaspers NG, Bootsma D, Hoeijmakers JH. Mutational analysis of the human nucleotide excision repair gene ERCC1. Nucleic Acids Res. 1996b;24:3370–3380. doi: 10.1093/nar/24.17.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijbers AM, Vader PCV, Snoek JW, Raams A, Jaspers NGJ, Kleijer WJ. Homozygous R788W point mutation in the XPF gene of a patient with xerodermapigmentosum and late-onset neurologic disease. J. Invest. Dermatol. 1998a;110:832–836. doi: 10.1046/j.1523-1747.1998.00171.x. [DOI] [PubMed] [Google Scholar]
- Sijbers AM, van Voorst Vader PC, Snoek JW, Raams A, Jaspers NG, Kleijer WJ. Homozygous R788W point mutation in the XPF gene of a patient with xeroderma pigmentosum and late-onset neurologic disease. J. Invest. Dermatol. 1998b;110:832–836. doi: 10.1046/j.1523-1747.1998.00171.x. [DOI] [PubMed] [Google Scholar]
- Smith DH, Fiehn AM, Fogh L, Christensen IJ, Hansen TP, Stenvang J, Nielsen HJ, Nielsen KV, Hasselby JP, Brunner N, et al. Measuring ERCC1 protein expression in cancer specimens: validation of a novel antibody. Sci. Rep. 2014;4:4313. doi: 10.1038/srep04313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, Scharer OD. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Orelli B, Madireddy A, Niedernhofer LJ, Scharer OD. Multiple DNA binding domains mediate the function of the ERCC1–XPF protein in nucleotide excision repair. J. Biol. Chem. 2012;287:21846–21855. doi: 10.1074/jbc.M111.337899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Ng JMY, Masutani C, Iwai S, van der Spek PJ, Eker APM, Hanaoka F, Bootsma D, Hoeijmakers JHJ. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH, Brookman KW, Dillehay LE, Mooney CL, Carrano AV. Hyper-sensitivity to mutation and sister-chromatid-exchange induction in CHO cell mutants defective in incising DNA containing UV lesions. Somat. Cell Genet. 1982;8:759–773. doi: 10.1007/BF01543017. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Brookman KW, Weber CA, Salazar EP, Reardon JT, Sancar A, Deng ZM, Siciliano MJ. Molecular-cloning of the human nucleotide-excision-repair gene ERCC4. Proc. Natl. Acad. Sci. U. S. A. 1994;91:6855–6859. doi: 10.1073/pnas.91.15.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Shinkura R, Shinkura N, Alt FW. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol. Cell. Biol. 2004;24:1200–1205. doi: 10.1128/MCB.24.3.1200-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson AE, Bardwell AJ, Bardwell L, Tappe NJ, Friedberg EC. Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single-stranded-DNA endonuclease. Nature. 1993;362:860–862. doi: 10.1038/362860a0. [DOI] [PubMed] [Google Scholar]
- Tripsianes K, Folkers GE, Zheng C, Das D, Grinstead JS, Kaptein R, Boelens R. Analysis of the XPA and ssDNA-binding surfaces on the central domain of human ERCC1 reveals evidence for subfunctionalization. Nucleic Acids Res. 2007;35:5789–5798. doi: 10.1093/nar/gkm503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsodikov OV, Enzlin JH, Scharer OD, Ellenberger T. Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF–ERCC1. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11236–11241. doi: 10.1073/pnas.0504341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsodikov OV, Ivanov D, Orelli B, Staresincic L, Shoshani I, Oberman R, Scharer OD, Wagner G, Ellenberger T. Structural basis for the recruitment of ERCC1–XPF to nucleotide excision repair complexes by XPA. EMBO J. 2007;26:4768–4776. doi: 10.1038/sj.emboj.7601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duin M, de Wit J, Odijk H, Westerveld A, Yasui A, Koken MH, Hoeijmakers JH, Bootsma D. Molecular characterization of the human excision repair gene ERCC-1: cDNA cloning and amino acid homology with the yeast DNA repair gene RAD10. Cell. 1986;44:913–923. doi: 10.1016/0092-8674(86)90014-0. [DOI] [PubMed] [Google Scholar]
- van Duin M, Koken MHM, Van DEN, Ten DIJKE, Odijk H, Westerveld A, Bootsma D, Hoeijmakers JHJ. Genomic characterisation of the human DNA excision repair gene ERCC-1. Nucleic Acids Res. 1987;15:9195–9214. doi: 10.1093/nar/15.22.9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duin M, van Den Tol J, Hoeijmakers JH, Bootsma D, Rupp IP, Reynolds P, Prakash L, Prakash S. Conserved pattern of antisense overlapping transcription in the homologous human ERCC-1 and yeast RAD10 DNA repair gene regions. Mol. Cell. Biol. 1989;9:1794–1798. doi: 10.1128/mcb.9.4.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houten B, Gamper H, Holbrook SR, Hearst JE, Sancar A. Action mechanism of ABC excision nuclease on a DNA substrate containing a psoralen crosslink at a de-fined position. Proc. Natl. Acad. Sci. U. S. A. 1986;83:8077–8081. doi: 10.1073/pnas.83.21.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vuuren AJ, Appeldoorn E, Odijk H, Yasui A, Jaspers NGJ, Hoeijmakers JHJ. Evidence for a repair enzyme complex involving ERCC1, ERCC4, ERCC11 and the xeroderma pigmentosum group F proteins. EMBO J. 1993;12:3693–3701. doi: 10.1002/j.1460-2075.1993.tb06044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez KM. Targeting and processing of site-specific DNA interstrand crosslinks. Environ. Mol. Mutagen. 2010;51:527–539. doi: 10.1002/em.20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Seo HY, Robinson A, Sowa G, Bentley D, Taylor L, Studer R, Usas A, Huard J, Alber S, et al. Accelerated aging of intervertebral discs in a mouse model of progeria. J. Orthop. Res. 2010;28:1600–1607. doi: 10.1002/jor.21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- Wan B, Yin J, Horvath K, Sarkar J, Chen Y, Wu J, Wan K, Lu J, Gu P, Yu EY, et al. SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell Rep. 2013;4:861–869. doi: 10.1016/j.celrep.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZG, Wu XH, Friedberg EC. Nucleotide excision repair of DNA in cell-free extracts of the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 1993;90:4907–4911. doi: 10.1073/pnas.90.11.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda G, Donker I, de Wit J, Morreau H, Janssens R, Vissers CJ, Nigg A, van Steeg H, Bootsma D, Hoeijmakers JHJ. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol. 1997;7:427–439. doi: 10.1016/s0960-9822(06)00190-4. [DOI] [PubMed] [Google Scholar]
- Westerveld A, Hoeijmakers JH, van Duin M, de Wit J, Odijk H, Pastink A, Wood RD, Bootsma D. Molecular cloning of a human DNA repair gene. Nature. 1984;310:425–429. doi: 10.1038/310425a0. [DOI] [PubMed] [Google Scholar]
- Wood RD. Mammalian nucleotide excision repair proteins and interstrand crosslink repair. Environ. Mol. Mutagen. 2010;51:520–526. doi: 10.1002/em.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood R, Burki HJ. Repair capability and the cellular age response for killing and mutation induction after UV. Mutat. Res. 1982;95:505–514. doi: 10.1016/0027-5107(82)90281-0. [DOI] [PubMed] [Google Scholar]
- Wood RD, De Veciana M, Presson-Tincknell B. Postirradiation properties of a UV-sensitive variant of CHO. Photochem. Photobiol. 1982;36:169–174. doi: 10.1111/j.1751-1097.1982.tb04359.x. [DOI] [PubMed] [Google Scholar]
- Wood RD, Burki HJ, Hughes M, Poley A. Radiation-induced lethality and mutation in a repair-deficient CHO cell line. Int. J. Radiat. Biol. 1983;43:207–213. doi: 10.1080/09553008314550241. [DOI] [PubMed] [Google Scholar]
- Wu Y, Mitchell TR, Zhu XD. Human XPF controls TRF2 and telomere length maintenance through distinctive mechanisms. Mech. Ageing Dev. 2008;129:602–610. doi: 10.1016/j.mad.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, III, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]