Abstract
Plant WRKY transcription factors are known to regulate various biotic and abiotic stress responses. In this study we identified a total of 30 putative WRKY unigenes in a transcriptome dataset of the Chinese wild Hazel, Corylus heterophylla, a species that is noted for its cold tolerance. Thirteen full-length of these ChWRKY genes were cloned and found to encode complete protein sequences, and they were divided into three groups, based on the number of WRKY domains and the pattern of zinc finger structures. Representatives of each of the groups, Unigene25835 (group I), Unigene37641 (group II) and Unigene20441 (group III), were transiently expressed as fusion proteins with yellow fluorescent fusion protein in Nicotiana benthamiana, where they were observed to accumulate in the nucleus, in accordance with their predicted roles as transcriptional activators. An analysis of the expression patterns of all 30 WRKY genes revealed differences in transcript abundance profiles following exposure to cold, drought and high salinity conditions. Among the stress-inducible genes, 23 were up-regulated by all three abiotic stresses and the WRKY genes collectively exhibited four different patterns of expression in flower buds during the overwintering period from November to April. The organ/tissue related expression analysis showed that 18 WRKY genes were highly expressed in stem but only 2 (Unigene9262 and Unigene43101) were greatest in male anthotaxies. The expression of Unigene37641, a member of the group II WRKY genes, was substantially up-regulated by cold, drought and salinity treatments, and its overexpression in Arabidopsis thaliana resulted in better seedling growth, compared with wild type plants, under cold treatment conditions. The transgenic lines also had exhibited higher soluble protein content, superoxide dismutase and peroxidase activiety and lower levels of malondialdehyde, which collectively suggets that Unigene37641 expression promotes cold tolerance.
Introduction
Members of the Corylus genus, commonly referred to as hazel, are economically and ecologically import plants in many parts of the world. Hazelnut kernels are rich in unsaturated fats, vitamin E, arginine, glutamic acid, and aspartic acid [1–3], and the nuts are incorporated in butters, pastes, confectionary spreads, flours, and are also widely used in the chocolate industry [4, 5]. In addition, the compound taxol (paclitaxel), a drug that is used in cancer therapy, is found in many hazelnut species [6, 7]. The major countries producing hazel for commercial purposes are Turkey, Italy, America, Azerbaijan, Georgia, and China [8–10], and currently more than 70% of the world’s production originates from the Black Sea region. More than 4 million acres of natural hazel grow in the northeastern and northwestern regions of China, with an annual yield of more than 23,000 tons [11–12].
The hazel species C. heterophylla Fisch is widely distributed in northern China where, for centuries, its nuts have been harvested for oil extraction and as a food source [3]. It is an economically important species, especially in the northeast region, and its production accounts for nearly three quarters of the total output of the Chinese domestic market [13]. In addition to its economic value, C. heterophylla also plays a key role in water and soil conservation and in the ecological balance of certain types of forests [14]. Some of the qualities that make C. heterophylla especially attractive as a hazelnut crop species include high productivity, early maturity, resistance to Eastern Filbert Blight (EFB), a fungus that is found in the common hazel, C. avellana [15, 16] and cold hardiness. Indeed, C. heterophylla has been shown to tolerate temperatures as low as -48°C, while C. avellana cannot [3, 17]. These desirable traits also make it a potentially important genetic resource for selection and breeding [18], and a C. heterophylla × C. avellana interspecific crossing project has been initiated in China and Korea [3].
The marked cold stress tolerance of C. heterophylla has provoked interest in identifying the key genes involved in the associated cold stress responses and the underlying signaling pathways [19]. Abiotic factors such as cold, drought, and high salinity, can cause severe damage in plants, resulting in major losses in crop yield and quality worldwide [20, 21]. In order to survive, plants to such abiotic stresses include numerous and complex biochemical and physiological changes [22]. The perception, signal transduction and molecular response mechanism of the external stimuli has been analyzed at the transcriptional level in many plant species [23, 24]. In the previous study, we used high resolution RNA-sequencing (RNA-seq) technology to elucidate the mechanisms of cold tolerance used by C. heterophylla, through a transcriptional profiling of four stages of floral buds, including non-cold acclimation (NA), cold acclimation (CA), midwinter (MW), and de-acclimation (DA) samples [25]. RNA-seq analysis has already proved useful for studies of C. avellana and C. mandshurica [2, 9]. We focused in particular on the transcriptional regulation of the cold stress responses and the role of transcription factors.
One of the largest families of transcription factors in plants is the WRKY family [26–28], members of which are known to modulate stress response [29–31]. A defining feature of this family, the WRKY domain, is a highly conserved stretch of approximately 60 amino acids with the highly conserved amino acid sequence WRKYGQK at the N-terminus, as well as the zinc finger structures C2H2 (CX4–5CX22–23HXH) or C2HC (CX7CX23HXC) at the C-terminus [32–35]. WRKY proteins can be classified into three groups, depending on the number of WRKY domains and the pattern of the zinc finger structures [27, 36]. In general, group I contains two WRKY domains, whereas the other two groups have only one domain. Group III contains a C2HC zinc finger motif that is distinct from the C2H2 motif present in group II [32]. To date, WRKY proteins have been shown to act as activators or repressors of developmental processes [27], such as trichome development [37], senescence [38, 39], embryogenesis [40], and seed development [41]. In addition, they have been shown to play a role in defense against biotic and abiotic stresses, including bacterial [42] and viral pathogens [43], wounding [44] drought, high salinity, cold and freezing [45]. However, the role of WRKY transcription factors in response to abiotic stresses is less well understood than the association with biotic stress factors [36, 46].
Since the first description of a WRKY protein, SPF1 from sweet potato (Ipomoea batatas) [47], large numbers of WRKY proteins have been identified from numerous plant species [48–55]. However, to date no WRKY transcription factor has been reported for any other Corylus species. In this study, we identified 30 members of the C. heterophylla WRKY transcription factor family, based on a transcriptome analysis of floral buds. We evaluated their expression in various tissues/organs under normal growing conditions and also following exposure to different abiotic stresses, including cold, drought and high salinity. In addition, we determined the subcellular localization of three members of the WRKY family and performed a functional assessment of one WRKY gene, Unigene37641, by overexpression in transgenic Arabidopsis thaliana plants.
Materials and Methods
Ethics statement
All necessary permits for field sampling were obtained from the local forestry department and Chinese Academy of Forestry.
Plant material and stress treatments
C. heterophylla was obtained from Weichang, Chengde, China (41°58′ N, 117°40′ E). Floral buds were collected on the first day of each month from November 2011 to March 2012. For organ/tissue-specific expression analysis, the stem and male anthotaxy was also collected in December. C. heterophylla plants were grown in pots containing sand and turf peat (1:2 v/v) in controlled environment growth chambers (16 h light/8 h dark cycle; 100 μmol·m−2·s−1 light intensity; 25°C) for two months. For drought, salinity, and cold stress treatments, seedlings were subjected to 25% (w/v) Polyethylene glycol (PEG) 6000, 400 mM NaCl, and 4°C conditions, respectively, as previously described by Wang et al [56]. Untreated seedlings were grown under the same environmental. Leaf samples were collected at 2 h, 4 h, 8 h and 24 h after treatment, frozen in liquid nitrogen and stored at −80°C prior to RNA extraction.
Cloning and sequence analysis of ChWRKY genes
Total RNA from C. heterophylla leaves, floral buds, stems and male anthotaxies were extracted using an RNA Extract Kit (Aidlab, Beijing, China). Based on the functional annotation of the C. heterophylla transcriptome (National Center for Biotechnology Information SRA database, accession number: SRX529300) [25], the assembled sequences were subjected to BLASTX (http://blast.ncbi.nlm.nih.gov/) analysis against the A. thaliana protein database at NCBI. Based on this analysis, a total of 30 candidate sequences containing WRKY domains were selected with an E-value less than 10−5. Primers for RACE were designed based on the WRKY sequences (S1 Table) and the RACE was conducted using the BD SMART RACE cDNA Amplification Kit (Clontech, Mountain View, USA) according to the manufacturer's instructions [57]. The products from the RACE PCR were ligated into the pMD-18T vector (TaKaRa, Dalian, China), which was transformed into competent Escherichia coli DH5α cells. After blue-white colony screening, positive clones were identified, and sent to Beijing Genomics Institution (BGI) for sequencing. Sequences of the PtrWRKY and AtWRKY proteins were downloaded from the Phytozome v10.2 website (http://www.phytozome.net/poplar) and the Arabidopsis genome TAIR 9.0 website (http://www.Arabidopsis.org/index.jsp), respectively. The phylogenetic trees were constructed using MEGA4.1 (http://www.megasoftware.net/mega.html) by employing the maximum likelihood (ML) for full-length proteins and neighbour-joining (NJ) method for conserved WRKY domains with 1,000 bootstrap replicates [58, 59]. The alignments of conserved WRKY domains in each subclass were output using DNAman. The WRKY domains were predicted using the MEME (http://meme.nbcr.net/meme/cgi-bin/meme.cgi) [29, 60]. Protein subcellular localization was predicted by Euk-mPLoc 2.0 software (http://www.csbio.sjtu.edu.cn/bioinf/euk-multi-2/).
Quantitative Real-time PCR (qRT-PCR) analysis
First strand cDNA was synthesized from total RNA using a First Strand Synthesis system (Invitrogen, USA) according to the manufacturer’s instructions. To analyze the expression levels of the WRKY transcription factors, qRT-PCR reactions were performed using the Bio-Rad CFX96 Real-Time PCR System (BIO-RAD, USA) and the SYBR Green qPCR Mix (Takara, Japan) [61]. qRT-PCR reactions were performed in a total volume of 20 μl and cycling conditions were 95°C for 30 s, followed by 39 cycles of 95°C/10 s, 60°C/15 s, 72°C/30 s, followed by a melting curve analysis [27]. The ChActin gene was used as an internal control [25] and the expression levels of WRKY genes were calculated using the 2−ΔΔCt formula [62]. The primer pairs were designed using Primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) and listed in S2 Table. Each reaction was performed with three biological replicates.
Subcellular localization assay
The sequences corresponding to the ORFs without stop codons of Unigene25835, Unigene37641 and Unigene20441 were inserted into the pEarlyGate101 vector (ABRC stock DB3-683) to produce 35S::ChWRKY-YFP constructs using the Gateway cloning system (Invitrogen, USA). For subcellular localization analysis, transient expression of Nicotiana benthamiana lower leaf epidermal cells was performed as previously described [63] with some modifications. Plants were cultivated under short-day conditions (8 h light/ 16 h dark). When the Agrobacterium culture reached the stationary growth phase at 28°C with agitation, cells were pelleted and resuspended in infiltration buffer (100 μM acetosyringone in 10 mM MgCl2). Fluorescence was observed using a LSM 510 confocal laser scanning microscope (Carl Zeiss, Oberkochen, Germany).
Generation of transgenic A. thaliana plants
The sequence corresponding to the Unigene37641 ORF was cloned and inserted into the pBI121 vector using the primer pair: 5´-ACTAGTATGTCTGATGAACA—TAG-3´ and 5´-GGTACCTGGCTCTTGTTTAAAG-3´. The resulting pBI121- Unigene37641 vector, which contained the CaMV 35S promoter to drive expression of Unigene37641, was transformed into A. thaliana using the floral dip method [64]. Semi-quantitative RT-PCR was used to determine the gene expression level in T2 transgenic lines. The wild type (WT) and transgenic seedlings were cultured on MS medium for 1 week, followed by the cold stress treatment (4°C for 6 h per day) for 2 weeks, as previously described by Li et al [57].
Soluble protein, superoxide dismutase, peroxidase and malondialdehyde measurements
Six-week-old WT and T2 transgenic lines were transferred to a cold-chamber and maintained at 4°C under light for 12 h. Soluble protein content was determined by the coomassie brillant blue G-250 method [65]. Superoxide dismutase (SOD) was calculated as previously described by Giannopolitis et al [66] and peroxidase (POD) activity assats were performed using the guaiacol method [67]. The malondialdehyde (MDA) content was determined using the thiobarbituric acid method [29].
Results
Cloning and sequence analysis of C. heterophylla WRKY genes
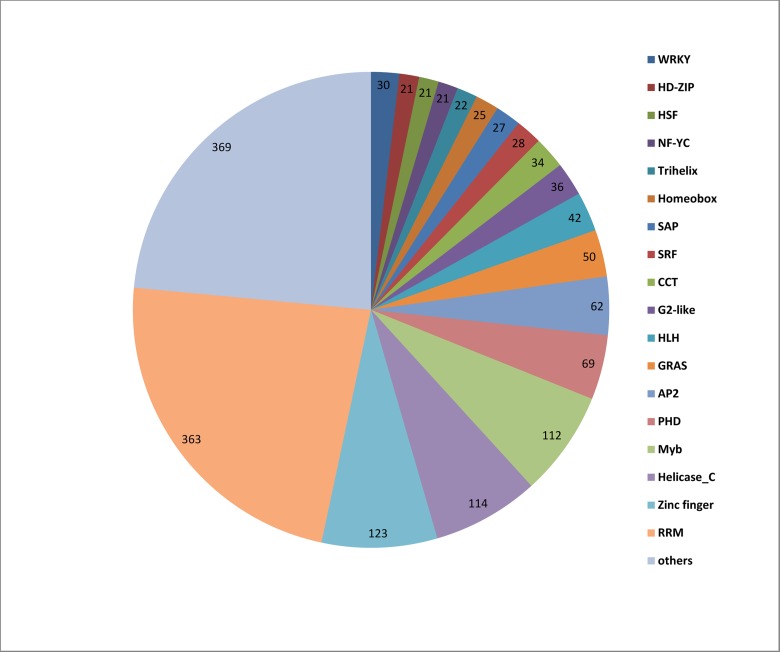
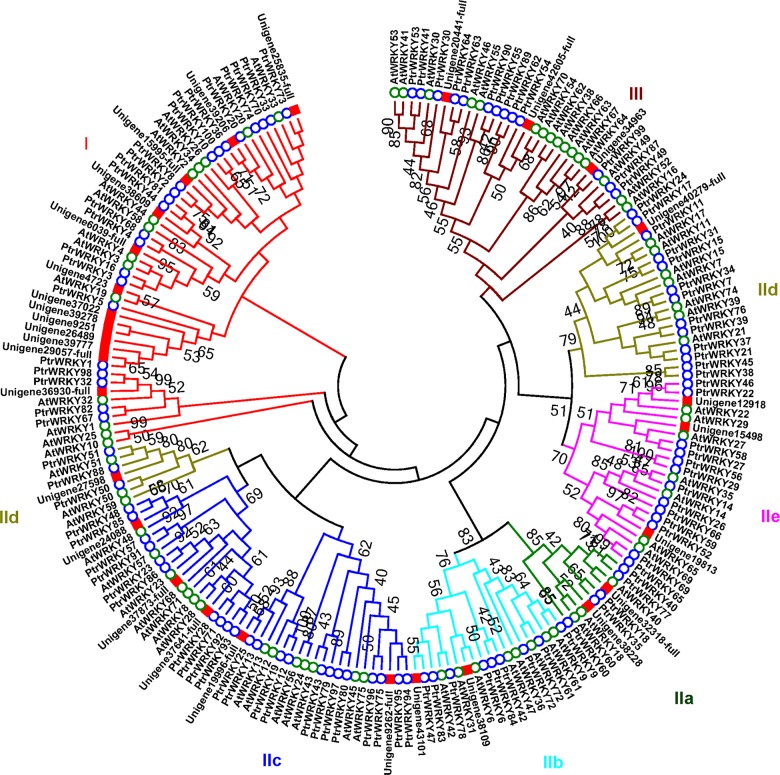
Following an analysis of the C. heterophylla floral bud transcriptome data set, we identified a total of 1,569 putative transcription factors and classified them into 64 families (Fig 1). Among them, 30 candidate genes encoding a WRKY domain were found and, using the Rapid Amplification of cDNA ends (RACE) technique, 13 full length genes with a complete open reading frame (ORF) were obtained (Table 1). Domain prediction clearly indicated that these proteins contained the conserved WRKY domain and zinc finger structure (Fig 2). To determine the phylogenetic relationships among the C. heterophylla WRKY proteins, an uprooted tree of all putative 30 ChWRKY proteins, 100 PtrWRKYs from Populus trichocarpa, and 72 AtWRKRs from A. thaliana was built using MEGA 4.1 (Fig 3). Based on the number of WRKY domains and the pattern of the zinc finger structures, the 30 corresponding WRKY proteins of C. heterophylla were divided into I, II and III groups. Group I contained 13 WRKY proteins, group III contained only 3 members with the specific zinc finger motif C2HC, while group II was further classified into five subgroups (IIa-e). Moreover, the phylogenetic tree was also constructed based on conserved WRKY domains. As shown in S1 Fig, group I contained sequences with a C-terminal WRKY domain or an N-terminal WRKY domain, and these sequences aligned within two different clusters, I-CT and I-NT, respectively. Of the thirty putative WRKY genes, fourteen belonged to group II, and one (Unigene37641) was chosen for further analysis. The full length cDNA of Unigene37641 is 1,342 bp (S2 Fig), including a predicted 963 bp ORF, which encodes a 320 amino acids polypeptide with a relative molecular mass of 35.06 kDa. The Unigene37641 protein contains a conserved WRKY domain and a zinc finger motif (C-X4-C-X23-H-X1-H), and a BLASTX analysis of the A. thaliana WRKY proteins in NCBI database revealed that AtWRKY28 is the most closely related protein to Unigene37641. The characteristics of other candidate WRKYs are provided in S3 Table.
Fig 1. Number of unique transcripts in annotated as transcription factors and the associated transcription factor families in the C. heterophylla Fisch transcriptome dataset.
Table 1. Characteristics of Corylus heterophylla Fisch WRKY genes.
| Unigene ID | Arabidopsis ortholog | E-value | cDNA length/ amino acids length | WRKY domain | Zinc finger motif | Subgroup |
|---|---|---|---|---|---|---|
| Unigene15995 | AtWRKY2 | 0.00E+00 | 2241/746 | WRKYGQK | C-X4-C-X23-H-X1-H | Group I |
| Unigene6039 | AtWRKY3 | 1.00E-131 | 1170/389 | WRKYGQK | C-X4-C-X23-H-X1-H | Group I |
| Unigene36930 | AtWRKY32 | 8.00E-134 | 1515/504 | WRKYGQK | C-X4-C-X23-H-X1-H | Group I |
| Unigene25835 | AtWRKY33 | 4.00E-116 | 1797/598 | WRKYGQK | C-X4-C-X23-H-X1-H | Group I |
| Unigene42605 | AtWRKY49 | 3.00E-14 | 1005/334 | WRKYGQK | C-X4-C-X22-H-X1-H | Group I |
| Unigene32318 | AtWRKY40 | 3.00E-105 | 954/317 | WRKYGQK | C-X5-C-X23-H-X1-H | Group II-a |
| Unigene19996 | AtWRKY13 | 2.00E-63 | 717/238 | WRKYGQK | C-X4-C-X23-H-X1-H | Group II-c |
| Unigene37873 | AtWRKY23 | 9.00E-70 | 957/318 | WRKYGQK | C-X4-C-X23-H-X1-H | Group II-c |
| Unigene37641 | AtWRKY28 | 6.00E-77 | 963/320 | WRKYGQK | C-X4-C-X23-H-X1-H | Group II-c |
| Unigene9262 | AtWRKY75 | 1.00E-55 | 540/179 | WRKYGKK | C-X4-C-X23-H-X1-H | Group II-c |
| Unigene29057 | AtWRKY11 | 1.00E-134 | 1035/344 | WRKYGQK | C-X5-C-X23-H-X1-H | Group II-d |
| Unigene40279 | AtWRKY11 | 2.00E-100 | 816/271 | WRKYGQK | C-X5-C-X23-H-X1-H | Group II-d |
| Unigene20441 | AtWRKY53 | 2.00E-52 | 1086/361 | WRKYGQK | C-X7-C-X23-H-X1-C | Group III |
Fig 2. Domain prediction of thirteen WRKY protein sequences.

The domain prediction, of thirteen C. heterophylla WRKY protein sequences and was performed using MEME software, which generated a letter logo to represent the WRKY domain and the zinc finger motif. The height of the letters in the y-axis represents the degree of conservation and relative frequency of each amino acid at that position.
Fig 3. Phylogenetic analyses of WRKY proteins from C. heterophylla, A. thaliana, and P. trichocarpa.
The phylogenetic tree of 30 putative C. heterophylla WRKYs, 72 AtWRKYs and 100 PtrWRKYs was constructed using MEGA 4.1, with a maximum likelihood (ML) method and 1,000 bootstrap replicates.
Expression analyses of WRKY genes during the overwintering
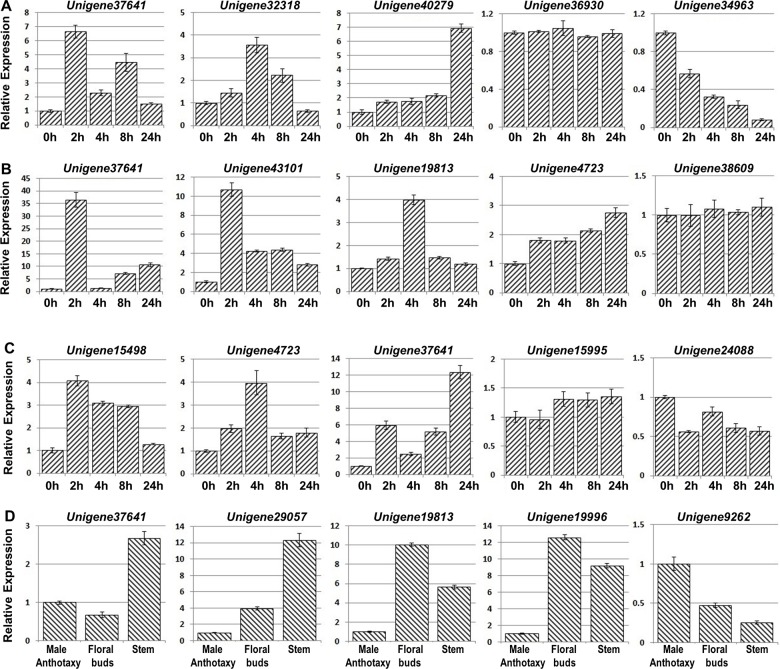
We evaluated the expression patterns of the 30 candidate WRKY genes in floral buds from C. heterophylla plants grown under normal conditions during the overwintering period, from November to April, by quantitative PCR (qRT-PCR) analysis. The WRKY genes were divided into four types according to their expression patterns (Fig 4 and S3 Fig): Type I (Unigene37641, Unigene43101, Unigene9251, Unigene4723, Unigene15995, Unigene40279, Unigene12918, Unigene37873, Unigene34963, Unigene38109, Unigene36930 and Unigene9262) showed the highest expression in November, which then declined until April; Type II (Unigene19996, Unigene15498, Unigene42605, Unigene37022, Unigene39206, Unigene39278, Unigene39777, Unigene25835, Unigene19813, Unigene38228, Unigene38609 and Unigene24088) showed an initial increase in expression followed by a subsequent decrease; Type III (Unigene32318, Unigene26489 and Unigene27598) low expression levels initially, followed by a significant increase in April; and Type IV (Unigene29057, Unigene20441 and Unigene6039) showed a decline after a general trend of increasing expression.
Fig 4. qRT-PCR analysis of WRKY gene expression during the overwintering.
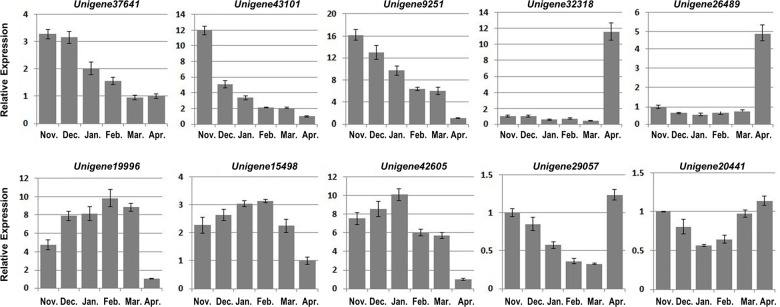
C. heterophylla Fisch floral buds were collected each month from 2011 November to 2012 April. The ChActin gene was used as an internal control for qRT-PCR analysis. The relative expression (y-axis) was calculated using the 2−ΔΔCt formula. The mean value and standard error were obtained from three biological and three technical replicates.
Expression patterns of ChWRKYs under cold, drought and salinity stresses
To identify the potential functions of the 30 WRKY genes in response to external stimuli, their expression profiles were analyzed in C. heterophylla leaves following cold, drought and salinity treatments. At 4°C, the expression of Unigene37641, Unigene32318, Unigene9262 and Unigene39278 was up-regulated with a maximal increase in expression of 6.7, 3.6, 3.6 and 7.0 fold after 2, 4, 8 and 24h, respectively (Fig 5A and S4 Fig). Similarly, 28 of the WRKY genes were up-regulated and reached maximum expression levels from 2-24h after the onset of a treatment with NaCl (Fig 5C and S6 Fig), while a drought treatment, induced by the application of polyethylene glycol (PEG6000), resulted in the up-regulation of 21 WRKY genes, which peaked at 2h (Fig 5B and S5 Fig). The expression of Unigene36930, Unigene38609 and Unigene15995 did not change as a consequence of the cold, drought or salinity treatments (Fig 5), but the expression of Unigene34963 and Unigene24088 was down regulated by the cold and salinity treatment, respectively (Fig 5A and 5C). Of the 30 ChWRKY genes, 23 were up-regulated by all three abiotic stresses (S4 Table), including Unigene37641, which was selected for further functional analysis by over-expression in transgenic A. thaliana plants.
Fig 5. Expression patterns of WRKY genes after exposure to various abiotic stresses and in different organs/tissues.
The ChActin gene was used as an internal control for qRT-PCR. The y-axis represents relative expression, calculated using the 2−ΔΔCt formula. (A-C) Expression profiles of WRKY genes under cold (4°C), drought (25% PEG6000) and salinity (400mM NaCl) growth conditions, respectively. Leaf samples were collected at 2, 4, 8 and 24h, with the untreated seedlings grown and analyzed in parallel. The expression level values of the genes in untreated seedlings were set to 1.0. (D) Expression patterns of WRKY genes in different organs/tissues, including male anthotaxies, floral buds and stems, which were collected at the same stage. The experiments were repeated with at least three biological and three technical replicates, yielding consistent results.
Expression patterns of WRKY genes in different organs/tissues under cold growth conditions
The expression of all 30 WRKY genes was evaluated in the male anthotaxy, floral buds and stems (Fig 5D and S7 Fig). The expression levels of Unigene37641 and Unigene29057 in stems, and of Unigene19813 and Unigene19996 in floral buds, were 2.7, 12.4, 10.1 and 12.6 fold greater, respectively, than in the male anthotaxy (Fig 5D), while the expression of Unigene9262 in the male anthotaxy was 3.9 and 2.1 fold greater, respectively, than in stems and floral buds. The expression analysis also revealed considerable variation in WRKY gene expression among the three organs/tissues, with 18 genes being highly expressed in stems and 10 in floral buds. In contrast, only 2 WRKY genes (Unigene9262 and Unigene43101) were abundantly expressed in the male anthotaxy.
Unigene25835, Unigene37641 and Unigene20441 localized in the nucleus
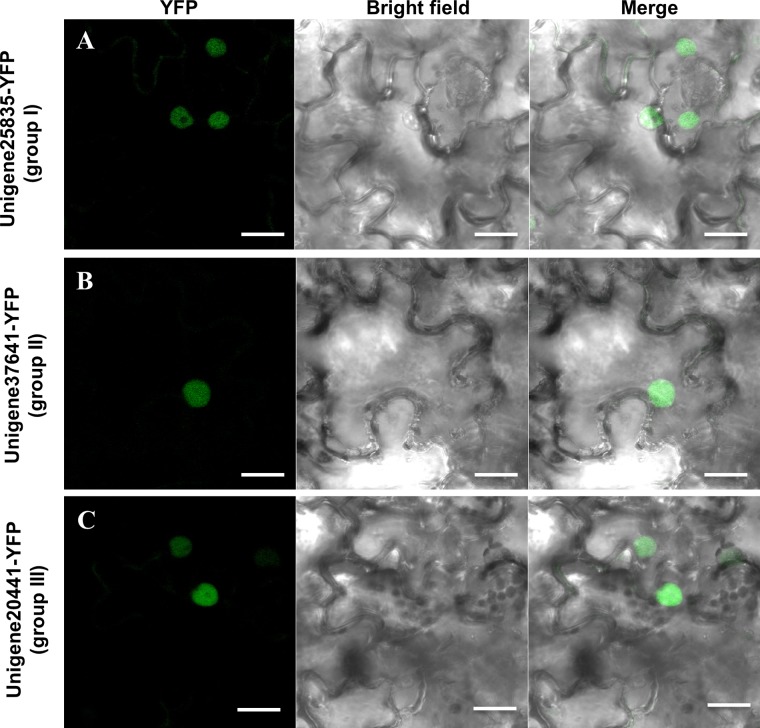
Computational analysis using the Euk-mPLoc software predicted that Unigene25835 (group I), Unigene37641 (group II) and Unigene20441 (group III) localize to the nucleus (S5 Table). To confirm their subcellular localization, we transiently expressed the proteins, each fused to the yellow fluorescent protein (YFP) reporter, in Nicotiana benthamiana leaf abaxial epidermal cells, under the control of the constitutive 35S promoter. As predicted, each of the fusion proteins was observed to exclusively accumulate in the nucleus of the epidermal cells (Fig 6), suggesting that Unigene25835, Unigene37641 and Unigene20441 are nuclear proteins, in accordance with their predicted function as transcription factors.
Fig 6. Subcellular localization of WRKY proteins.
Confocal images of Nicotiana benthamiana epidermal leaf cells expressing Group I Unigene25835 (A), Group II Unigene37641 (B) and Group III Unigene20441 (C) WRKY proteins fused to yellow fluorescent protein (YFP). Scale bar = 20 μm.
Overexpression of Unigene37641 enhanced cold tolerance in A. thaliana
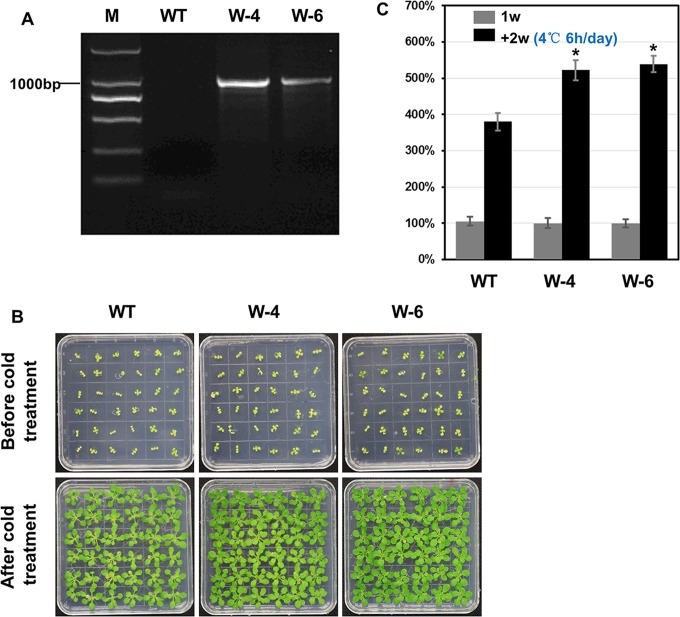
To investigate the function of Unigene37641, we overexpressed the gene in A. thaliana. RT-PCR analysis showed that two T2 transgenic lines (W-4 and W-6) had high levels of expression of the Unigene37641 gene, while expression was not detected in wild type plants (Fig 7A). After cold treatment for 2 weeks, seedlings of the two Unigene37641 overexpressing transgenic lines showed more vigorous growth than the wild type (WT) seedlings and this was confirmed by a quantitative analysis of the average fresh weight (Fig 7B and 7C). A cold tolerance test further showed higher levels of soluble protein and greater SOD and POD activities in the transgenic lines than in WT plants (S8A–S8C Fig). Moreover, MDA levels, which provide a measure of the degree of damage to cell membranes caused by lipid peroxidation, were substantially lower in the transgenic lines that in WT plants following cold stress (S8D Fig). These results suggest that overexpression of Unigene37641 enhanced cold stress tolerance in the transgenic A. thaliana.
Fig 7. Unigene37641 overexpression enhanced cold tolerance in transgenic A. thaliana.
(A) Expression level of Unigene37641 in T2 Arabidopsis transgenic lines (W-4 and W-6) and wild type (WT) plants using RT-PCR. (B) The WT and T2 plants were cultured on MS medium for 1 week were shown in top row. Seedlings were grown at 4°C (6 h per day) for 2 weeks were shown in bottom row. (C) Comparison of seedling fresh weight after cold treatment. The mean value of fresh weights and the standard errors were calculated based on three replicates.
Discussion
Plant WRKY transcription factors comprise a superfamily involved in the regulation of a variety of development processes and stress responses [57, 68]. However, compared with the considerable progress that has been made in understanding their role in biotic stresses, less is known about their function in the context of abiotic stresses [29]. Numerous WRKY transcription factors have been identified in various plant species, such as A. thaliana, Oryza sativa (rice), P. trichocarpa (poplar), Glycine max (soybean) and Pinus monticola (pine) [69–72] and there is considerable interest in investigating their role in responses to factors such as drought, high salinity and cold temperature, in the context of effects on crop yield and quality [73]. In this study, we investigated the composition and potential functions of the WRKY gene family of the hazelnut tree C. heterophylla Fisch, an economically and ecologically species with several traits that make it valuable as an agricultural commodity, one of which being its hardiness in cold temperatures. A total of 30 putative WRKY unigenes were identified, of which 13 members were cloned to obtain full length sequences with complete ORFs, and they were all found to have conserved structural features, including WRKY domains and zinc finger regions (Fig 2).
A phylogenetic tree of WRKY proteins from C. heterophylla, A. thaliana, and P. trichocarpa was constructed to examine their evolutionary relationships (Fig 3). According to the report by Eulgem et al [32], all these corresponding proteins could be classified into three main groups (I, II and III) based on the number of WRKY domains and the type of zinc-finger motif (Table 1). Interestingly, the phylogenetic analyses demonstrated that Unigene42605 sequence aligned within clade III. However, this protein has a C2H2 zinc finger structure that is different from the typical C2HC sequence that is typical of type of group III (Fig 3 and S9 Fig). Based on the phylogenetic tree of conserved WRKY domains and alignment analyses, Unigene42605, which contained a C-X4-C-X22-H-X1-H motif, should be assigned to group I (S1 and S9 Figs). Furthermore, Unigene29057 clustered in subgroup IId based on the conserved WRKY domains (S1 Fig). This finding was inconsistent with the results shown in Fig 3, where this protein was classified in group I. The sequence alignments demonstrated that Unigene29057 contained only one WRKY domain with a C-X5-C-X23-H-X1-H zinc finger, implying that this member should belong to group IId (S9 Fig). A phylogenetic tree combining WRKYs from different species has the potential to not only elucidate the evolutionary relationships of the proteins [70], but also to allow predictions of the functions of the C. heterophylla proteins based on the functional clades of the orthologous proteins, since close homologs can share expression profiles and functions [74]. For example, AtWRKY28 and PtrWRKY28 aligned within group IIc (Fig 3), members of which are associated with responses to abiotic stresses [70, 75]. Thus, we can infer that Unigene37641 might have similar roles since it clustered with the same group. Subcellular localization studies showed that Unigene25835-YFP (group I), Unigene37641-YFP (group II) and Unigene20441-YFP (group III) fusion proteins accumulated in the nucleus of N. benthamiana leaf epidermal cells following transient expression (Fig 6), which is in accordance with their computationally predicted localization (S5 Table) and previous studies of other species [76].
We next examined the expression patterns of the WRKY transcription factors in the floral buds of overwintering C. heterophylla. It has previously been determined that the floral buds undergo four developmental stages during winter: NA (non-cold acclimation), CA (cold acclimation), MW (midwinter) and DA (de-acclimation) [25]. The expression data showed that several WRKY genes, such as Unigene37641, Unigene43101 and Unigene9251, were highly expressed in the early CA stage in November, with a subsequent decrease in expression (Fig 4). These WRKY genes may be responsible for activating the expression of downstream target genes to enhance cold tolerance of the floral buds during the winter season. Unigene19996, Unigene15498 and Unigene42605 showed a different expression pattern, in that they showed the highest transcript levels at the MW stage, suggesting an involvement in later cold acclimation. In contrast, Unigene32318, Unigene26489 and Unigene27598 were not upregulated from November to March but showed a substantial increase in expression in April (Fig 4 and S3 Fig), suggesting functions other than responses to low temperatures [25]. These genes may be induced by other environmental cues, such as short photoperiods, and contribute to developmental processes [27] and/or flowering time regulation [77]. Based on the expression profiles, we hypothesize that Unigene32318, Unigene26489 and Unigene27598 may be involved in floral bud development, since C. heterophylla floral buds germinate in April. Finally, the expression of Unigene29057 and Unigene20441 showed an initial decline followed by an increasing trend (Fig 4); however, since individual WRKY genes have been found to function in both environmental stimuli and plant development [27, 73], it may be that these two genes are involved in both cold acclimation and flower development.
To assess the potential influence of other environmental factors on the observed expression patterns of the WRKY genes, we induced other abiotic stresses on the plants using an artificial climate chamber. qRT-PCT analyses revealed that 24 WRKY genes were significantly up-regulated and 2 down-regulated by cold stress (S4 Table). Interestingly, the expression of Unigene32318 and Unigene26489 was substantially induced by the cold treatment, peaking after 4h (Fig 5A and S4 Fig), even though their expression was not observed to increase from November to March under overwintering conditions (Fig 4). This suggests that their expression is induced by multiple factors, potentially involving several signaling pathways, implying a complex regulatory network [29]. Of the 30 WRKY genes identified here, the expression of 28 changed following the drought and salt treatments (S4 Table), indicating a general correlation between drought and salinity stress responses, and the existence of crosstalk between the respective signal transduction pathways [71, 78]. As shown in Fig 5, some of the WRKY genes showed an extremely rapid response to abiotic stress: the expression of Unigene37641, Unigene43101 and Unigene15498 peaked at 2h after the cold, drought and salinity treatments, respectively. We note that an increasing number of reports describe rapid expression of WRKY transcription factors in association with stress tolerance [32, 73]. When the expression profiles of the 30 WRKY genes were evaluated in three different organs/tissues, 18 were most highly expressed in the stem and only 2 genes were strongly expressed in the male anthotaxy. This difference in organ/tissue specific expression suggests functional divergence of the respective genes [30].
We observed that the expression of Unigene37641 was significantly up-regulated by all three stresses and we selected this gene for functional analysis by overexpression in A. thaliana. Several previous studies have reported that overexpression of WRKY genes in A. thaliana can enhance its tolerance to various abiotic stresses [36], one example being the overexpression of soybean GmWRKY21, which led to enhanced cold tolerance [71]. Another example came from the overexpression of the rice OsWRKY45 and OsWRK72 genes, which conferred drought and salt tolerance to transgenic A. thaliana plants [79, 80]. Compared with WT plants, transgenic A. thaliana overexpressing Unigene37641 showed no obvious phenotypic differences. However, the seedlings were less susceptible to cold stress (Fig 7B), which is in accordance with a previous report in which grapevine VpWRKY2 was overexpressed in A. thaliana, resulting in enhanced cold stress tolerance [57]. Unigene37641 overexpressing plants also showed levels of higher soluble protein, SOD and POD activities, and lower MDA levles compared to WT controls following the cold treantment (S8 Fig). This suggests that Unigene37641 may help protect the plant by a mechanism that includes reducing membrane damage that would result in increased MDA production [81]. Several studies have shown similar expression patterns of putative WRKY genes and orthologs from A. thaliana [74], and overexpression of AtWRKY28 in A. thaliana was recently shown to enhance tolerance to various stresses, including drought, salinity, oxalic acid and fungal pathogens, suggesting diverse regulatory functions [75, 82, 83]. Given that, of the 30 C. heterophylla WRKY genes, Unigene37641 is the most closely related to AtWRKY28, we propose that Unigene37641 may exhibit similar expression patterns and possibly have similar functions [74].
In conclusion, the regulatory mechanisms of WRKY proteins involved in stress tolerance are complex and further studies are needed to elucidate their functions. This current investigation of WRKY genes expression in C. heterophylla also provides a platform for further exploring the function of WRKY genes in other species and suggests candidate genes for enhancing biotic and abiotic stress tolerance in crops.
Supporting Information
A phylogenetic tree of conserved WRKY domains, built using MEGA 4.1 and employing the neighbour-joining (NJ) method with 1,000 bootstrap replicates. Group I was clustered into two groups, I-CT and I-NT, based on the C-terminal WRKY domain and N-terminal WRKY domains, respectively.
(TIF)
The cysteine and the histidine residues of the zinc-finger motif are boxed and the shaded area represents the WRKY domain.
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Six-week-old wild type (WT) and T2 transgenic lines (W-4, W-6) were held at 4°C for 24 h. (A) Soluble protein content in WT and T2 transgenic leaves exposed to cold. (B) Superoxide dismutase activity. (C) Peroxidase activity. (D) Malondialdehyde content. The mean values and standard errors were derived from three experimental replicates.
(TIF)
Based on the features of their WRKY domains, the corresponding proteins from C. heterophylla and P. trichocarpa were divided into three groups. Groups II was further classified into five subgroups (IIa, IIb, IIc, IId, IIe). The WRKYGQK domains are indicated with red boxes and the zinc-finger motif sequences are indicated with red triangles.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank the Key Laboratory of Tree Breeding and Cultivation of State Forestry Administration of China for providing basic experimental conditions. We would like to thank PlantScribe (www.plantscribe.com) for carefully editing on this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Support was provided by The Special Fund for Basic Scientific Research Business of Central Public Research Institutes (Grant No. RIF2013-10 http://rif.caf.ac.cn/), the Special Fund from State Forestry Administration in the Public Interest (Grant No. 201304710 http://www.forestry.gov.cn/), and Beijing Municipal Natural Science Foundation (Grant No. 6144030 http://www.bjnsf.org/).
References
- 1. Özdemir M, Seyhan FG, Bakan AK, I̊lter S, Özay G, et al. Analysis of internal browning of roasted hazelnuts. Food Chem. 2001; 73: 191–196. [Google Scholar]
- 2. Ma H, Lu ZQ, Liu BB, Qiu Q, Liu JQ. Transcriptome analyses of a Chinese hazelnut species Corylus mandshurica . BMC Plant Biol. 2013; 13:152 10.1186/1471-2229-13-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mehlenbacher SA. Hazelnuts. In: Moore JN, Ballington JR, editors. Genetic resources in temperate fruit and nut crops. Acta Horticulturae. 1991; 290: 789–836. [Google Scholar]
- 4. Ozdemir F, Akinci I. Physical and nutritional properties of four major commercial Turkish hazelnut varieties. J Food Eng. 2004; 63: 341–347. [Google Scholar]
- 5. Fallico B, Arena E, Zappala M. Roasting of hazelnuts. Role of oil in colour development and hydroxymethylfurfural formation. Food Chem. 2003; 81: 569–573. [Google Scholar]
- 6. Kumar S, Mahdi H, Bryant C, Shah JP, Garg G, et al. Clinical trials and progress with paclitaxel in ovarian cancer. Inter J Women’s Health. 2010; 2:411–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luo F, Fei X, Tang F, Li X. Simultaneous determination of Paclitaxel in hazelnut by HPLC-MS/MS. For Res. 2011; 24:779–783. [Google Scholar]
- 8. Erdogan V, Mehlenbacher SA. Phylogenetic relationships of Corylus species (Betulaceae) based on nuclear ribosomal DNA ITS region and chloroplast matK gene sequences. Syst Bot. 2000; 25: 727–737. [Google Scholar]
- 9. Rowley ER, Fox SE, Bryant DW, Sullivan CM, Priest HD, et al. Assembly and characterization of the European hazelnut 'Jefferson’ transcriptome. Crop Sci. 2012; 52:2679–2686. [Google Scholar]
- 10. Gökirmak T, Mehlenbacher SA, Bassil NV. Characterization of European hazelnut (Corylus avellana) cultivars using SSR markers. Genet Resour Crop Ev. 2009; 56:147–172. [Google Scholar]
- 11.FAO website. Available: http://www.fao.org/docrep/003/X4484E/x4484e03.htm
- 12. Zhang Y, Li F, Tao R, Li Z, Liang Y. An investigation of wild Corylus resource at Changbai Mountains. J Jilin Agri Sci. 2007; 32:56–57. [Google Scholar]
- 13. Liu J, Cheng Y, Liu C, Zhang C, Wang Z. Temporal changes of disodium fluorescein transport in hazelnut during fruit development stage. Sci Hortic Amsterdam. 2013; 150: 348–353. [Google Scholar]
- 14. Liu H. Exploring the utilization of Corylus. Farm Prod Proc. 2010; 1:24–25. [Google Scholar]
- 15. Chen HL, Mehlenbacher SA, Smith DC. Hazelnut accessions provide new sources of resistance to Eastern Filbert Blight. Hortscience. 2007; 42:466–469. [Google Scholar]
- 16. Molnar TJ, Capik J, Zhao S, Zhang N. First report of Eastern Filbert Blight on Corylus avellana 'Gasaway’ and 'VR20-11’ caused by Anisogramma anomala in New Jersey. Plant Dis. 2010; 94:1265–1265. [DOI] [PubMed] [Google Scholar]
- 17. Peng L, Wang M, Liang W, Xie M, Li D. A study on cold resistance for filbert genus (Corylus L.) plants. J Jilin Fores Univ. 1994; 3:166–170. [Google Scholar]
- 18. Ni B, Ni W, Xu X, Wang X. Hazel breeding research. Forest By-Product and Speciality in China. 2010; 106:29–31. [Google Scholar]
- 19. Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Biol. 1999; 50: 571–599. [DOI] [PubMed] [Google Scholar]
- 20. Singh KB, Foley RC, Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002; 5:430–436. [DOI] [PubMed] [Google Scholar]
- 21. Bohnert HJ, Nelson DE, Jensen RG. Adaptations to environmental stresses. Plant Cell. 1995; 7: 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant Journal. 2002; 31:279–292. [DOI] [PubMed] [Google Scholar]
- 23. Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol and Plant Mol Biol. 2000; 51:463–499. [DOI] [PubMed] [Google Scholar]
- 24. Wang HH, Hao JJ, Chen XJ, Hao ZN, Wang X, et al. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol. 2007; 65: 799–815. [DOI] [PubMed] [Google Scholar]
- 25. Chen X, Zhang J, Liu QZ, Guo W, Zhao TT, et al. Transcriptome Sequencing and Identification of Cold Tolerance Genes in Hardy Corylus Species (C. heterophylla Fisch) Floral Buds. PLoS One. 2013; 9:e108604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol. 2004; 7:491–498. [DOI] [PubMed] [Google Scholar]
- 27. Qin Z, Lv HJ, Zhu XL, Meng C, Quan TY, et al. Ectopic expression of a wheat WRKY transcription factor gene TaWRKY71-1results in hyponastic leaves in Arabidopsis thaliana . PLoS One. 2013; 8:e63033 10.1371/journal.pone.0063033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yin GJ, Xu HL, Xiao SY, Qin YJ, Li YX, et al. The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 2013; 13:148 10.1186/1471-2229-13-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang C, Deng PY, Chen LL, Wang XT, Ma H, et al. A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS One. 2013; 8:e65120 10.1371/journal.pone.0065120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramamoorthy R, Jiang SY, Kumar N, Venkatesh PN, Ramachandran S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008; 49:865–879. 10.1093/pcp/pcn061 [DOI] [PubMed] [Google Scholar]
- 31. Reddy AS, Ali GS, Celesnik H, Day IS. Coping with stresses: roles of calcium-and calcium/calmodulin-regulated gene expression. Plant Cell. 2011; 23: 2010–2032. 10.1105/tpc.111.084988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000; 5:199–206. [DOI] [PubMed] [Google Scholar]
- 33. Kalde M, Barth M, Somssich IE, Lippok B. Members of the Arabidopsis WRKY Group III Transcription Factors Are Part of Different Plant Defense Signaling Pathways. Mol Plant Microbe In. 2003; 16:295–305. [DOI] [PubMed] [Google Scholar]
- 34. Xie Z, Zhang ZL, Zou XL, Huang J, Ruas P, et al. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells, Plant Physiol. 2005; 137:176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Song Y, Ai CR, Jing SJ, Yu DQ. Research progress on functional analysis of rice WRKY genes. Rice Science. 2010; 17:60–72. [Google Scholar]
- 36. Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010; 15:247–258. 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- 37. Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002; 14:1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hinderhofer K, Zentgraf U. Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta. 2001; 213:469–473. [DOI] [PubMed] [Google Scholar]
- 39. Robatzek S, Somssich IE. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defencerelated processes. Plant J. 2001; 28:123–133. [DOI] [PubMed] [Google Scholar]
- 40. Lagacé M, Matton DP. Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense . Planta. 2004; 219:185–189. [DOI] [PubMed] [Google Scholar]
- 41. Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASEgene, are regulators of seed size in Arabidopsis . Proc Natl Acad Sci USA. 2005; 48:17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ishihama N, Yoshioka H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr Opin Plant Biol. 2012; 15:431–437. 10.1016/j.pbi.2012.02.003 [DOI] [PubMed] [Google Scholar]
- 43. Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007; 10:366–371. [DOI] [PubMed] [Google Scholar]
- 44. Hara K, Yagi M, Kusano T, Sano H. Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol Gen Genet. 2000; 263:30–37. [DOI] [PubMed] [Google Scholar]
- 45. Huang T, Duman JG. Cloning and characterization of a thermal hysteresis (antifreeze) protein with DNAbinding activity from winter bittersweet night shade, Solanum dulcamara . Plant Mol Biol. 2002; 48:339–350. [DOI] [PubMed] [Google Scholar]
- 46. Niu C, Wei W, Zhou Q, Tian A, Hao Y, et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012; 35: 1156–1170. 10.1111/j.1365-3040.2012.02480.x [DOI] [PubMed] [Google Scholar]
- 47. Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 59 upstream regions of genes coding for sporamin and b-amylase from sweet potato. Mol Gen Genet. 1994; 244:563–571. [DOI] [PubMed] [Google Scholar]
- 48. Marchive C, Léon C, Kappel C, Coutos-Thévenot P, Corio-Costet MF, et al. Over-Expression of VvWRKY1 in grapevines induces expression of jasmonic acid pathway-related genes and confers higher tolerance to the downy mildew. PloS One. 2013; 8: e54185 10.1371/journal.pone.0054185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Luo X, Bai X, Sun X, Zhu D, Liu B, et al. Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J Exp Bot. 2013; 64: 2155–2169. 10.1093/jxb/ert073 [DOI] [PubMed] [Google Scholar]
- 50. Proietti S, Bertini L, Van der Ent S, Leon-Reyes A, Pieterse CM, et al. Cross activity of orthologous WRKY transcription factors in wheat and Arabidopsis . J Exp Bot. 2011; 62: 1975–1990. 10.1093/jxb/erq396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tao Z, Kou Y, Liu H, Li X, Xiao J, et al. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J Exp Bot. 2011; 62: 4863–4874. 10.1093/jxb/err144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dang FF, Wang YN, Yu L, Eulgem T, Lai Y, et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013; 36:757–774. 10.1111/pce.12011 [DOI] [PubMed] [Google Scholar]
- 53. Xiong X, James VA, Zhang H, Altpeter F. Constitutive expression of the barley HvWRKY38 transcription factor enhances drought tolerance in turf and forage grass (Paspalum notatum Flugge). Mol Breeding. 2010; 25: 419–432. [Google Scholar]
- 54. Ren X, Chen Z, Liu Y, Zhang H, Zhang M, et al. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010; 63: 417–429. 10.1111/j.1365-313X.2010.04248.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li H, Gao Y, Xu H, Dai Y, Deng D, et al. ZmWRKY33, a WRKY maize transcription factor conferring enhanced salt stress tolerances in Arabidopsis . Plant Growth Regul. 2013; 70:207–216. [Google Scholar]
- 56. Wang LQ, Wang C, Wang DY, Wang YC. Molecular characterization and transcript profiling of NAC genes in response to abiotic stress in Tamarix . Tree Genet Genomes. 2014; 10:157–171. [Google Scholar]
- 57. Li H, Xu Y, Xiao Y, Zhu ZG, Xie XQ, et al. Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild Vitis pseudoreticulata . Planta Med. 2010; 232:1325–1337. [DOI] [PubMed] [Google Scholar]
- 58. Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003; 52:696–704. [DOI] [PubMed] [Google Scholar]
- 59. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007; 24:1596–1599. [DOI] [PubMed] [Google Scholar]
- 60. Wilkinson MD, Castells-Brooke N, Shewry PR. Diversity of sequences encoded by the Gsp-1 genes in wheat and other grass species. J Cereal Sci. 2013; 57: 1–9. [Google Scholar]
- 61. Zhang YC, Zhang J, Song TT, Li JY, Tian J, et al. Low medium pH value enhances anthocyanin accumulation in Malus crabapple leaves. PloS One. 2014; 9:e97904 10.1371/journal.pone.0097904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 63. Zheng H, Camacho L, Wee E, Batoko H, Legen J, et al. A Rab-EGTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell. 2005; 17: 2020–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 1998; 16:735–743. [DOI] [PubMed] [Google Scholar]
- 65. Gao JF. Experimental guidance for plant physiology. 1st ed Beijing: Higher Education Press; 2006. [Google Scholar]
- 66. Giannopolitis CN, Ries SK. Superoxide dismutases I. occurrence in higher plant. Plant Physiol. 1977; 59:309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang ZL, Qu WJ. Experimental guidance for plant physiology. 3st ed Beijing: Higher Education Press; 2003. [Google Scholar]
- 68. Zhang Y, Feng JC. Identification and characterization of the grape WRKY family. Biomed Res Int. 2014; 787680 10.1155/2014/787680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wu KL, Guo ZJ, Wang HH, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Research. 2005; 12:9–26. [DOI] [PubMed] [Google Scholar]
- 70. Jiang YZ, Duan YJ, Yin J, Ye SL, Zhu JR, et al. Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J Exp Bot. 2014; 65:6629–6644. 10.1093/jxb/eru381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, et al. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J. 2008; 6:486–503. 10.1111/j.1467-7652.2008.00336.x [DOI] [PubMed] [Google Scholar]
- 72. Liu JJ, Ekramoddoullah AK. Identification and characterization of the WRKY transcription factor family in Pinus monticola . Genome. 2009; 52:77–88. 10.1139/G08-106 [DOI] [PubMed] [Google Scholar]
- 73. Chen LG, Song Y, Li SJ, Zhang LP, Zou CS, et al. The role of WRKY transcription factors in plant abiotic stresses. BBA-Gene Regul Mech. 2012; 1819:120–128. [DOI] [PubMed] [Google Scholar]
- 74. Ling J, Jiang WJ, Zhang Y, Yu HJ, Mao ZC, et al. Genome-wide analysis of WRKY gene family in Cucumis sativus . BMC Genomics. 2011; 12:471 10.1186/1471-2164-12-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Babitha KC, Ramu SV, Pruthvi V, Mahesh P, Nataraja KN, et al. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis . Transgenic Res. 2013; 22:327–341. 10.1007/s11248-012-9645-8 [DOI] [PubMed] [Google Scholar]
- 76. Lippok B, Birkenbihl RP, Rivory G, Brümmer J, Schmelzer E, et al. Expression of AtWRKY33 encoding a pathogenor PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol Plant Microbe Interact. 2007; 20:420–429. [DOI] [PubMed] [Google Scholar]
- 77. Luo X, Sun XL, Liu BH, Zhu D, Bai X, et al. Ectopic Expression of a WRKY Homolog from Glycine soja Alters Flowering Time in Arabidopsis . PLoS One. 2013; 8:e73295 10.1371/journal.pone.0073295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002; 31:279–292. [DOI] [PubMed] [Google Scholar]
- 79. Qiu YP, Yu DQ. Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis . Environ Exp Bot. 2009; 65:35–47. [Google Scholar]
- 80. Song Y, Chen LG, Zhang LP, Yu DQ. Overexpression of OsWRKY72 gene interferes in the ABA signal and auxin transport pathway of Arabidopsis . Biosci. 2010; 35: 459–471. [DOI] [PubMed] [Google Scholar]
- 81. Sathiyaraj G, Lee OR, Parvin S, Khorolragchaa A, Kim YJ, et al. Transcript profiling of antioxidant genes during biotic and abiotic stresses in Panax ginseng C. A. Meyer. Mol Biol Rep. 2011; 38: 2761–2769. 10.1007/s11033-010-0421-7 [DOI] [PubMed] [Google Scholar]
- 82. Chen XT, Liu J, Lin GF, Wang AR, Wang ZH, et al. Overexpression of AtWRKY28 and AtWRKY75 in Arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum . Plant Cell Rep. 2013; 32:1589–1599. 10.1007/s00299-013-1469-3 [DOI] [PubMed] [Google Scholar]
- 83. Wu LT, Zhong GM, Wang JM, Li XF, Song X, et al. Arabidopsis WRKY28 transcription factor is required for resistance to necrotrophic pathogen, Botrytis cinerea . AFR J Microbiol Res. 2011; 5:5481–5488. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A phylogenetic tree of conserved WRKY domains, built using MEGA 4.1 and employing the neighbour-joining (NJ) method with 1,000 bootstrap replicates. Group I was clustered into two groups, I-CT and I-NT, based on the C-terminal WRKY domain and N-terminal WRKY domains, respectively.
(TIF)
The cysteine and the histidine residues of the zinc-finger motif are boxed and the shaded area represents the WRKY domain.
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Six-week-old wild type (WT) and T2 transgenic lines (W-4, W-6) were held at 4°C for 24 h. (A) Soluble protein content in WT and T2 transgenic leaves exposed to cold. (B) Superoxide dismutase activity. (C) Peroxidase activity. (D) Malondialdehyde content. The mean values and standard errors were derived from three experimental replicates.
(TIF)
Based on the features of their WRKY domains, the corresponding proteins from C. heterophylla and P. trichocarpa were divided into three groups. Groups II was further classified into five subgroups (IIa, IIb, IIc, IId, IIe). The WRKYGQK domains are indicated with red boxes and the zinc-finger motif sequences are indicated with red triangles.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.