Abstract
CBF transcription factors, which play important roles in cold acclimation, regulate the expression of approximately 170 cold-responsive genes, termed the CBF regulon. Recent work by Park et al. showed that CBF regulon genes and other cold-responsive genes are regulated by a complex network that involves many early cold-induced transcription factors.
Keywords: cold stress, transcriptional regulatory network, CBF regulon, Arabidopsis
Roles of CBF transcription factors in cold acclimation
It is well known that exposing plants to low nonfreezing temperatures can increase their tolerance to freezing stress via a process called cold acclimation [1]. Cold acclimation has been detected in many temperate plants, such as winter wheat, barley, oats and rape [2]. During the last two decades, the mechanism of cold acclimation has been extensively studied. In Arabidopsis thaliana, researchers have identified CBF transcription factors, including CBF1, CBF2 and CBF3 (also known as DREB1b, DREB1c and DREB1a), that play important roles in cold acclimation [3]. CBF proteins, which are in the AP2/ERF (APETALA2/Ethylene-Responsive Factor) transcription factor family, are able to recognize and bind to the CRT/DRE (C-repeat/Dehydration Responsive Element) DNA regulatory motif in the promoters of many cold-responsive (COR) genes [4]. The three CBF genes are rapidly and transiently up-regulated after cold treatment, usually reaching their maximal expression levels after 1–3h at 4°C [3].
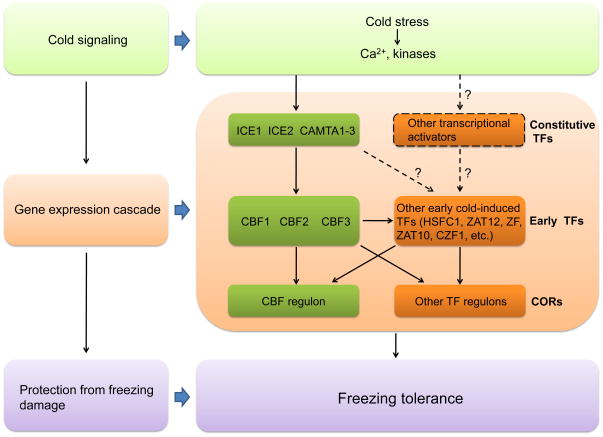
Current data show that the CBF genes are mainly regulated by ICE1, ICE2 and the three closely related CAMTA transcription factors, CAMTA1-3 [5–8] (Figure 1). Specifically, mutation of ice1 greatly affects the cold-induced gene expression of CBF2 and CBF3, but to a less degree affects the gene expression of CBF1 [5, 9]. Overexpression of ICE2 in transgenic plants mainly increases the expression of CBF1 [6]. Studies of single and double camta mutants indicated that the three CAMTA proteins are required for the regulation of all three CBFs, and it appears that CAMTA3 plays a more important role compared to CAMTA1 and CAMTA2 [7, 8]. These results suggest that although CBF1, CBF2 and CBF3 genes are physically linked in the genome and have similar biological functions, they are preferentially regulated by different upstream transcriptional activators.
Figure 1.
The complex network of cold stress regulated gene expression. Cold stress stimulates Ca2+ signaling and activates protein kinases. The protein kinases phosphorylate upstream transcription factors (TFs), resulting in the expression of early cold-responsive transcription factors (early TFs). The early TFs then activate the expression of downstream cold-responsive (COR) genes that confer freezing tolerance. Many of the COR genes are co-regulated by more than one early TFs, and some of the early TFs are regulated by CBFs and other early TFs. Therefore, the various TF regulons are interconnected.
The notion that CBF proteins contribute to cold acclimation is supported by the finding that constitutive overexpression of CBF1, CBF2 or CBF3 in transgenic plants induces the expression of many COR genes, termed the CBF regulon, and subsequently increases freezing tolerance even without prior cold exposure [3]. In contrast, knock-down of both CBF1 and CBF3 using antisense constructs downregulates the cold-induction of CBF regulon genes and decreases freezing tolerance [10]. How the proteins encoded by the CBF regulon impart freezing tolerance is not fully understood, but the cryoprotection properties of some COR proteins and metabolic alteration are thought to be important for freezing tolerance [2] (Figure 1).
Regulation of COR genes
Extensive studies have shown that cold acclimation largely relies on the regulation of COR gene expression. Analysis of gene expression datasets found more than 2000 COR genes. Approximately 1200 of these are robustly regulated by low temperature, about 170 of which belong to the CBF regulon [11]. Although genes regulated by CBF proteins are important for freezing tolerance, they only account for a small percentage of the COR genes, thus how the remaining COR genes are regulated needs to be investigated.
The first discovery concerning regulation of COR genes by non-CBF proteins was provided by Vogel et al. in 2005 [12], who found that the ZAT12 transcription factor regulates 24 COR genes. Recently, Park et al. [11] showed that there are 174 cold-induced genes encoding putative transcription factors, at least 17 of them are early cold-induced transcription factor genes with similar expression patterns as the CBFs. These genes include ZF, CZF1, RAV1, CZF2, MYB73, ZAT12, DOF1.10, ZAT10, HSFC1, DEAR1, MYB44, ERF5, CRF2, WRKY33, ERF6, CRF3 and RVE2. Through constitutive overexpression of 11 of these first-wave transcription factors in transgenic plants, the authors identified five (HSFC1, ZAT12, ZF, ZAT10 and CZF1) that confer the expression of 121, 67, 61, 54 and 51 COR genes, respectively, in the absence of cold treatment (Figure 1). This result clearly indicates that transcription factors other than CBF1, CBF2 and CBF3 are involved in the regulation of COR genes. Considering that only a small portion of cold-inducible transcription factors are tested, additional transcription factors are probably involved in the regulation of COR gene expression.
Co-regulation of the CBF regulon by other first-wave transcription factors
Since there are potentially many other regulons involved in COR gene expression in addition to the CBF regulon, an important question is whether and how much the different regulons may overlap. Vogel et al. [12] reported that 7 of the ZAT12 regulon genes are also members of the CBF2 regulon. Recently, Park et al. [11] found that 35 CBF2 regulon genes are also regulated by one or more of the other five early cold-induced transcription factors (Figure 1). Similarly, these other five first-wave transcription factors share some common downstream genes. These findings demonstrate that the low temperature regulatory network is highly interconnected, involving extensive crosstalk and co-regulation (co-regulation here refers to regulation of a gene by two or more transcription factors in an independent fashion). Interestingly, overexpression of CBF2, HSFC1 and CZF1 induces the gene expression of ZF, ZAT12 and ZAT10, respectively, suggesting that some of these first-wave transcription factors can regulate each other [11]. None of the CBF genes is regulated by the other five first-wave transcription factors, indicating that the effects of the other five first-wave transcription factors on the CBF regulon are due to co-regulation. In addition, in transgenic plants expressing a truncated, dominant negative form of CBF2, in which CBF-mediated gene expression is blocked, the cold-induced expression of CBF regulon genes is only partially affected [11]. This result supports that other cold-inducible transcription factors are involved in the co-regulation of the CBF regulon genes.
Concluding remarks and future perspectives
The recent study by Park et al. [11] clearly shows that although the first discovered CBF regulon is very important for cold acclimation, COR gene regulation involves many other cold-responsive transcription factor regulons, and there is extensive crosstalk and co-regulation of the different regulons. This study provides important new information indicating that cold regulated gene expression cascades are highly interconnected and are much more complex than previously thought. The interconnectedness suggests that cold responsive gene regulation in plants is very robust, but also implies that many more early cold-regulated transcription factors would need to be manipulated to achieve maximal improvements in freezing tolerance. Important questions remain to be answered. Firstly, only about one-third of the COR genes were identified as part of the regulons of CBFs and the other five first-wave transcription factors, thus how the remaining two-thirds of the COR genes are regulated is still unknown. One possibility is that these remaining COR genes are regulated by additional cold-responsive transcription factors. Alternatively, cold-triggered post-translational modifications may be required for the cold-responsive transcription factors to regulate the expression of the remaining COR genes. In this case, COR genes cannot be regulated by mere constitutive overexpression of the first-wave transcription factors at warm temperature, thereby many COR genes belonging to the regulons of the first-wave transcription factors would be missed in the study. Another possibility is that the expression of some of the COR genes may require the simultaneous presence of more than one early cold responsive transcription factors. Secondly, CBF gene expression is mainly regulated by ICE1, ICE2 and CAMTA1-3, and cold-induced expression of ZAT12 is reduced in camta3 mutant [7], thereby whether the other first-wave transcription factors are also regulated by these three upstream transcription factors or by other transcriptional activators needs to be addressed. Thirdly, constitutive overexpression of CBFs, ZAT12 or HSFC1 in transgenic plants is able to increase freezing tolerance, suggesting that the genes that are co-regulated by these first-wave transcription factors may play more important roles than other COR genes in cold acclimation. These co-regulated genes should be studied in more detail to deepen our understanding of the mechanism of cold acclimation.
Acknowledgments
This work was supported by U.S. NIGMS grant to J.-K.Z. (R01GM059138) and by the Chinese Academy of Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 2.Chinnusamy V, et al. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Medina J, et al. The CBFs: three arabidopsis transcription factors to cold acclimate. Plant sci. 2011;180:3–11. doi: 10.1016/j.plantsci.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 4.Stockinger EJ, et al. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinnusamy V, et al. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fursova OV, et al. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene. 2009;429:98–103. doi: 10.1016/j.gene.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Doherty CJ, et al. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell. 2009;21:972–984. doi: 10.1105/tpc.108.063958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y, et al. Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. The Plant J. 2013;75:364–376. doi: 10.1111/tpj.12205. [DOI] [PubMed] [Google Scholar]
- 9.Ding Y, et al. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev cell. 2015;32:278–289. doi: 10.1016/j.devcel.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 10.Novillo F, et al. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc Natl Acad Sci USA. 2007;104:21002–21007. doi: 10.1073/pnas.0705639105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park S, et al. Regulation of the Arabidopsis CBF regulon by a complex low temperature regulatory network. The Plant J. 2015;82:193–207. doi: 10.1111/tpj.12796. [DOI] [PubMed] [Google Scholar]
- 12.Vogel JT, et al. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]