Summary
CRISPR-based technologies have emerged as powerful tools to alter genomes and mark chromosomal loci, but an inexpensive method for generating large numbers of RNA guides for whole genome screening and labeling is lacking. Using a method that permits library construction from any source of DNA, we generated guide libraries that label repetitive loci or a single chromosomal locus in Xenopus egg extracts and show that a complex library can target the E. coli genome at high frequency.
Introduction
Recently, the potential for broad scale modification of specific genomic sequences has emerged with clustered regularly interspaced short palindromic repeats (CRISPR) technology. Based on an adaptive immune system in bacteria and archaea that protects against phage and other foreign nucleic acids (Wiedenheft et al., 2012), short CRISPR-derived RNAs bind to Cas (CRISPR-associated) proteins and direct them to degrade foreign DNA elements (Jinek et al., 2012). The CRISPR system of Streptococus pyogenes in particular has been harnessed as a genome-editing tool utilizing a chimeric synthetic guide RNA (sgRNA) and the Cas9 protein. These 2 components are sufficient to direct specific DNA binding and cleavage of DNA sequences complementary to the sgRNA (Jinek et al., 2013). Other recent innovations have used engineered versions of the Cas9 protein lacking nuclease activity (dCas9) fused to various protein domains as tools to repress or activate reporter gene expression in yeast and human cells (Gilbert et al., 2013; Qi et al., 2013). The use of fluorescent dCas9 fusions for labeling chromosome loci in cultured cells has also been described (Chen et al., 2013). However, this technique has been limited to single locus or repetitive loci targeted by a small number of designed guides. Although it is theoretically possible to expand labeling to larger regions or even whole vertebrate chromosomes by generating many thousands of guides, the complexity and cost of oligonucleotide synthesis makes this approach impractical for most laboratories. Similarly, genome-wide screening libraries are available for some well-studied organisms and have seen broad interest for loss-of-function screens, but generation of such libraries by oligonucleotide synthesis approaches is unlikely to be cost-effective for many other organisms otherwise amenable to CRISPR-mediated screens, or for which genome data is not yet available.
We set out to develop an approach for generating large numbers of diverse guide RNAs, both for our studies using a CRISPR-based system to label specific sequences on chromosomes in Xenopus egg extracts and, separately, applying the same technique to a prokaryotic genome to demonstrate its applicability in the generation of a guide library from an arbitrary source of DNA.
Design
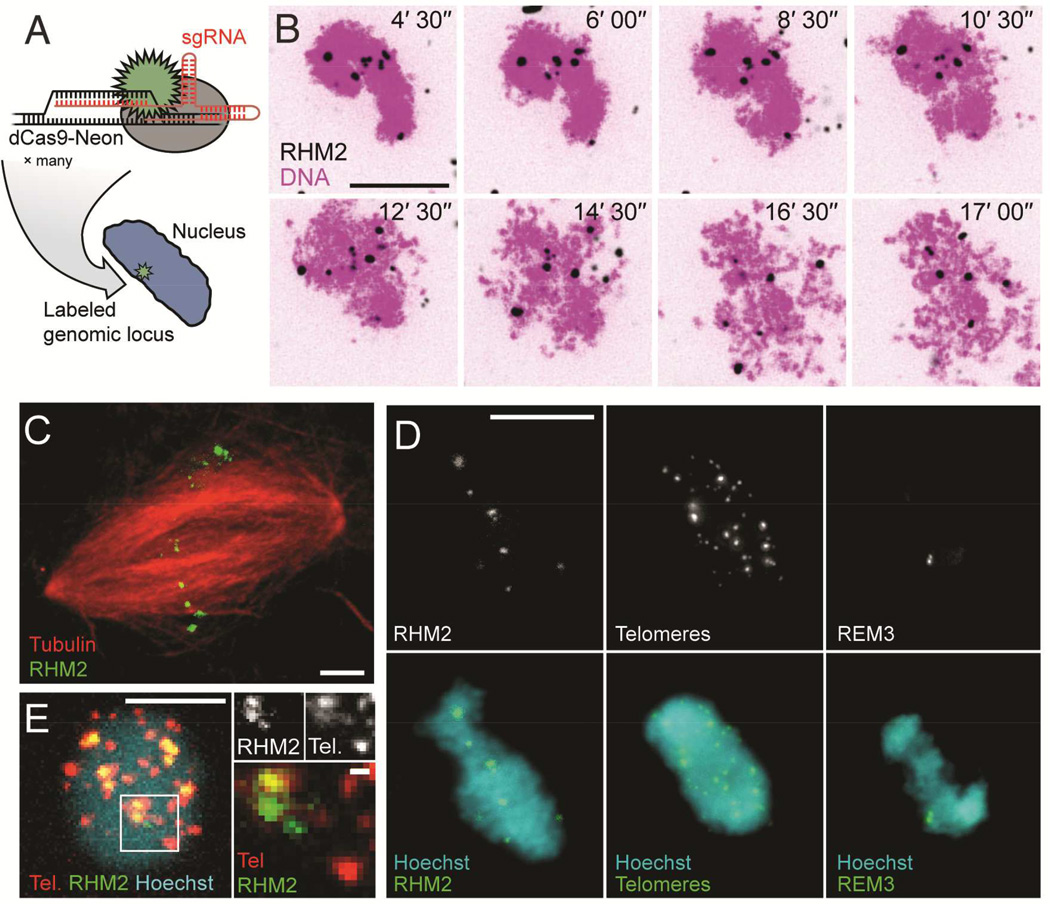
To label chromosomes in vitro, we expressed and purified recombinant nuclease deficient Cas9 (dCas9) fused to mNeonGreen, the brightest green/yellow fluorescent protein yet described (Shaner et al., 2013) (Figure 1A). The dCas9-Neon protein was complexed with 7 sgRNAs designed against the RHM2 745 base pair (bp) multiple-locus tandem repeat, present at a mean of ~2000 copies near the centromere of most Xenopus laevis chromosomes (Freeman and Rayburn, 2005; Meyerhof et al., 1983) (Table S1, S2). A big advantage of using Xenopus extract is that it can be biochemically manipulated and the cell cycle state controlled (Figure S1). We followed the dynamics of mitotic chromatid formation by time-lapse fluorescence microscopy by adding sperm nuclei to metaphase-arrested egg extract that has been ultracentrifuged to remove membranes. During this reaction, sperm chromatin remodels and individual chromatids resolve from one another. Upon addition of RHM2/dCas9-Neon probes, puncta formed in numbers in agreement with that expected from published in situ hybridization data (Freeman and Rayburn, 2005; Meyerhof et al., 1983); chromatids with distinct foci could be seen individualizing and separating from the chromosome mass within 10 minutes (Figure 1B, Movie S1, see also Figure S2). In crude extracts that support transit through the cell cycle, RHM2 labeling was maintained on mitotic chromosomes as the spindle formed, and probes were visible at the metaphase plate (Figure 1C). Two other classes of repeat were labeled in the same way with patterns in agreement with published data (Figure 1D) (Bassham et al., 1998; Hummel et al., 1984). Simultaneous dual-color labeling of two classes of repeats was also possible (Figure 1E).
Figure 1. Repetitive genomic loci can be visualized using dCas9-Neon in Xenopus egg extracts.
A: dCas9-Neon is programmed to label specific genomic loci by conjugation to an sgRNA molecule containing a complementary target sequence. See also Figure S1. B: dCas9-Neon programmed using RHM2 sgRNA (black) localizes rapidly to loci in sperm nuclei (Sytox Orange dye, magenta). Time (min) after imaging started is indicated in the top left of each image. See also Supplementary Movie 1 and Figure S2. C: Labeled RHM2 loci (green) are maintained following formation of a mitotic spindle (red). D: Three examples of repeat classes labeled on sperm nuclei in Xenopus egg extract (1n = 18). Left: RHM2 is a centromere-proximal locus on ~65% of chromosomes (Freeman and Rayburn, 2005). Middle: Telomere repeats target chromosome termini. Right: REM3 is reported to target a single centromere-proximal locus on chromosome 1, appearing here as two spots (Hummel et al., 1984). E: Left: Sperm nuclei driven into interphase in the presence of dCas9-tdTomato Telomere sgRNA and dCas9-Neon RHM2 sgRNA demonstrate simultaneous dual-color labeling (scale bar, 5 µm). Right: A subset of RHM2 and telomere loci appear to co-localize, while others do not (scale bars 10 µm, except magnification in panel E, 1 µm).
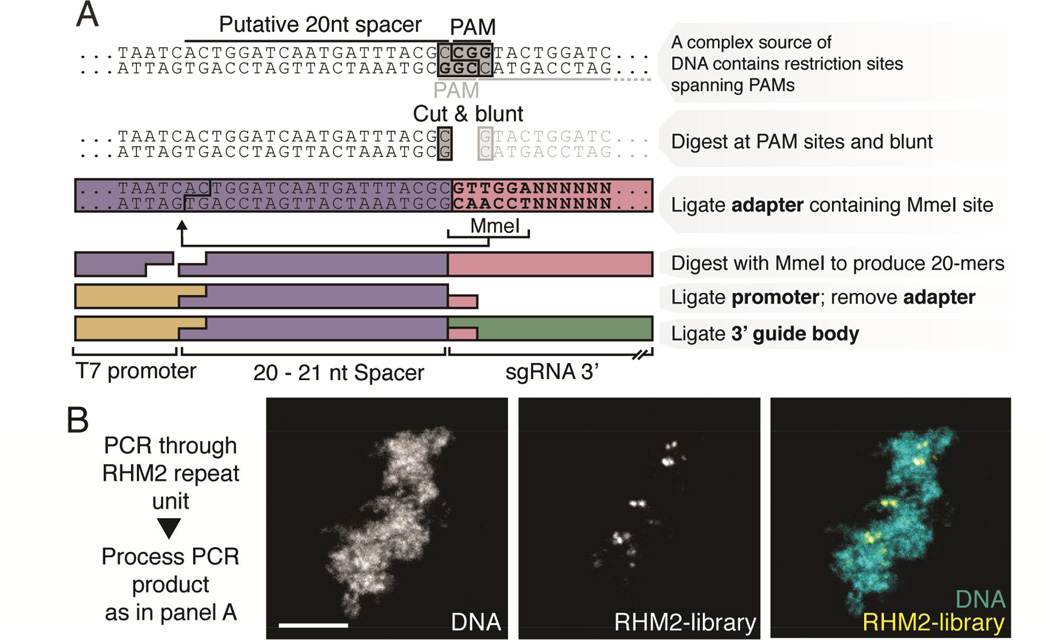
For labeling non-repetitive loci, we reasoned that potentially any DNA sequence could be enzymatically processed into a library of sgRNAs and used to tile along a chromosomal region. The constraints imposed by the S. pyogenes CRISPR system are that a targeted sequence must be approximately 20 nucleotides (nt) in length and immediately 5’ to a “PAM”, or protospacer adjacent motif consisting of an NAG or NGG triplet. We designed a strategy to extract PAM-proximal sequences by digesting input DNA with restriction enzymes targeting immediately 5’ to an NGG or NAG (see details in Materials and Methods). The resulting fragments are ligated to an adapter containing a recognition site for the restriction enzyme MmeI, which cuts 20–21 nt 5’. Finally, we removed the adapter and ligated the resulting fragments to a 5’ RNA polymerase promoter for in vitro transcription and a 93 nt 3’ sgRNA Cas9 hairpin (Figure 2A). We first evaluated the effectiveness of the digestion/ligation protocol in CRISPR imaging on the RHM2 repeat amplified by PCR and found that the probes gave similar labeling patterns as the traditionally designed guides (Figure 2B, compare to Figure 1B, D).
Figure 2. An enzymatically generated guide library can program dCas9-Neon labeling of a repetitive locus.
A: Outline of enzymatic library generation approach. B: dCas9-Neon programmed using an RHM2 repeat unit processed with this method localizes in a labeling pattern similar to that seen for RHM2 in Figure 1B and 1D (scale bar, 5 µm).
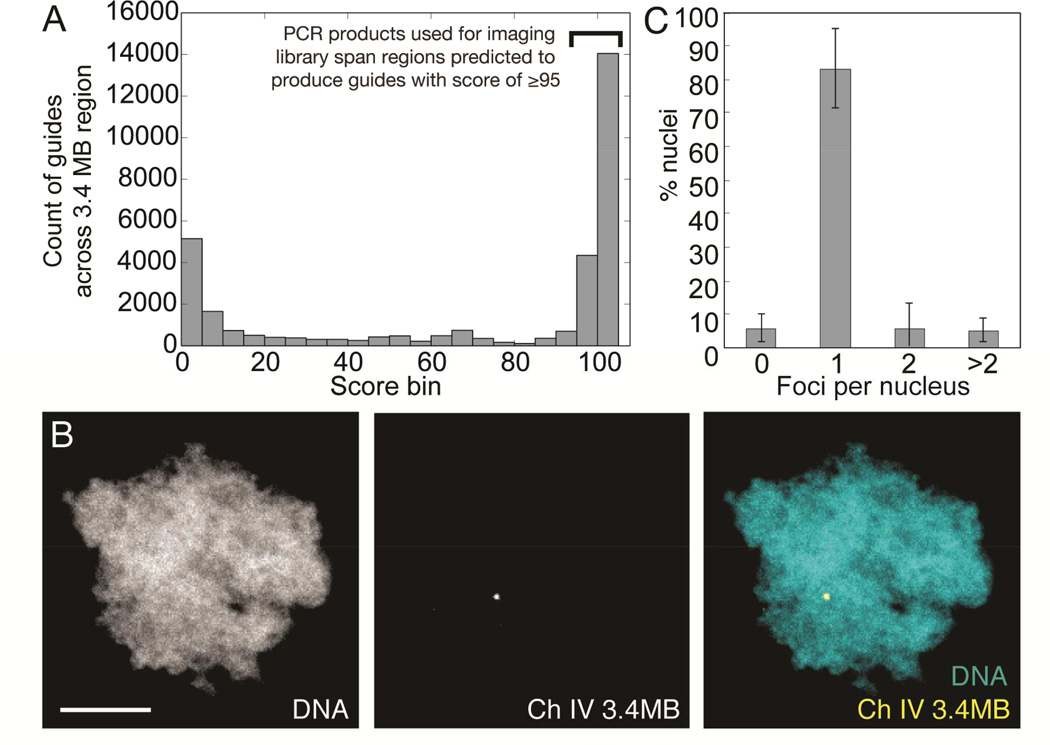
To label arbitrary, non-repetitive regions of the genome, we PCR-amplified specific subsequences within a 3.4 megabase (MB) region on chromosome 4 of the X. laevis genome. These subsequences represent 144 regions from 316 bp to 4088 bp in length that, when amplified, pooled and digested as described above are predicted to produce guides with minimal off-target effects (Table S3). The extent of off-target effects was predicted using a previously published scoring algorithm that determines the number and location of mismatches within guide target sequences when aligned to the entire genome (Hsu et al., 2013). A guide with no predicted off-target binding is scored as 100 in this scheme. We picked a threshold of 95, at which no perfect matches are found elsewhere in the genome and the closest matches differ at positions that would strongly impair guide recognition (Figure 3A, Supplemental Data S1–S3 and online repository; see Methods). We obtained 100 PCR products (see Figure S3), which we expected to yield 1,276 guides when all products were pooled and subjected to the digestion/ligation library protocol. After enzymatically processing the PCR products as outlined above, the final pooled library was transcribed in vitro using T7 RNA polymerase (see detailed protocol in Supplemental Files).
Figure 3. A single 3.4 MB locus can be labeled using an enzymatically generated guide library.
A: Specificity score distribution for all guides predicted to be generated by subjecting 3.4 MB region to procedure outlined in Figure 2A. Only sub-regions predicted to generate guides with a score of ≥95 were used as PCR templates for library construction. B: Processing of 100 PCR products (See Figure S3) spanning regions within a 3.4MB region of X. laevis chromosome 4 generates a single labeled spot in haploid sperm nuclei (scale bar, 5 µm). C: Count of fluorescent foci per sperm nucleus when incubated with 3.4 MB library. n = 3 experiments, 11–13 nuclei scored per experiment. Bars are ± standard deviation. See also Figure S3, Table S3, and Supplemental Data S1–S3.
Results
When incubated with dCas9-Neon in egg extract, the transcribed pooled PCR product library generated a single major spot in sperm nuclei (Figure 3B and C), demonstrating that this method provides an innovative, relatively inexpensive, and effective approach for live whole-chromosome labeling.
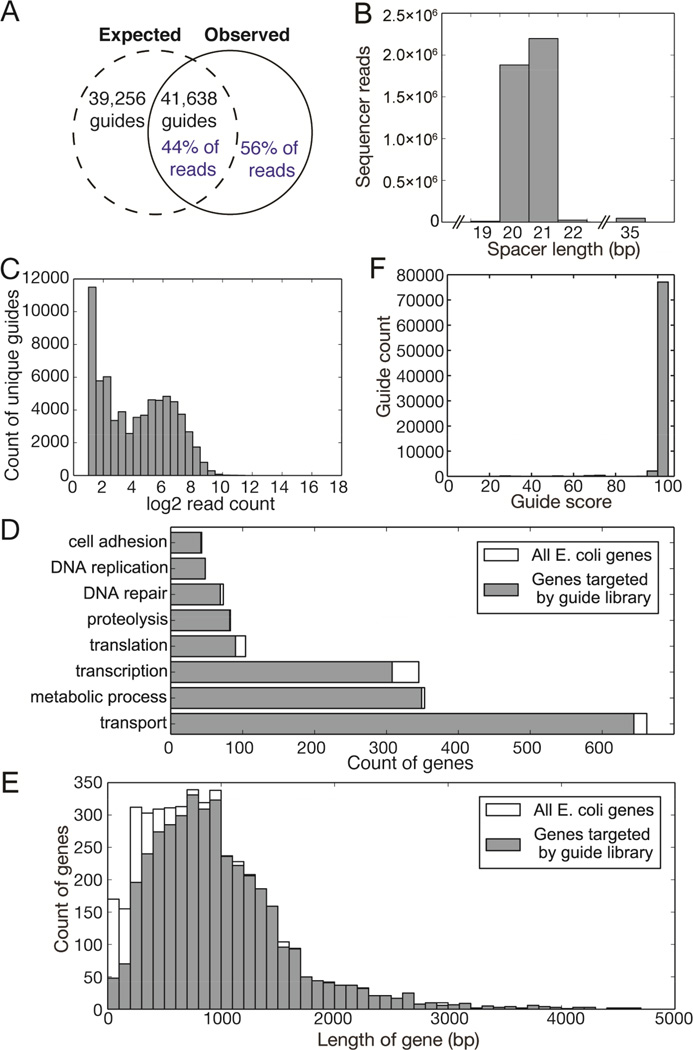
Having shown that the molecular approach to generating a library using the digestion/ligation protocol is possible, we explored its utility in making large, complex libraries suitable for use in genome-wide screens by CRISPR-mediated mutagenesis. In order to simplify analysis of the resulting library, we chose to use the well-characterized E. coli genome as a template. We extracted genomic DNA from a cloning strain of E. coli (XL1-Blue) and subjected it to the digestion/ligation protocol detailed above in parallel duplicates. Using publicly available E. coli genome sequence data, we calculated that 80,894 guides could theoretically be generated using this approach (Figure 4A). High-throughput sequencing of the library revealed 37,854 guides, at mean incidence of one guide for every 112 nt in the E. coli genome, representing ~44% of the total material sequenced. Of the remaining 56%, 45% of the total material consisted of guides shifted by 1–3 bases 3’ relative to PAMs, likely due to promiscuous activity of Mung-bean nuclease used to blunt fragments. The guides were otherwise consistent with the intended design, containing a T7 promoter followed by a 20–21 nt variable region (Figure 4B) and terminating with a 93 nt invariant region necessary for Cas9 binding.
Figure 4. A complex guide library targeting sequences within the E. coli genome.
A: Comparison of theoretical maximum number of guides generated by E. coli genome digestion with guides identified by sequencing (black text) and of sequencing reads that represent expected guides versus those reads that do not correctly target E. coli PAM-adjacent 20mers (blue text). B: Length distribution of variable spacers (region between T7 promoter and sgRNA guide body) in library as determined by high-throughput sequencing. C: Distribution of abundance of unique guides within library. D: Coverage of selected GO-term gene groups by library sgRNAs compared to the total number of genes annotated by those GO terms. E: Analysis of genes targeted by guides in sequenced library as binned by gene length. F: In silico analysis of guide specificity as predicted to be produced by digestion/ligation of E. coli genomic DNA. A score of 100 indicates no predicted off-target effects.
An ideal library is one that exhibits high complexity and is composed of equal numbers of molecules representing each unique guide sequence. However, libraries are subject to deviation from this ideal, due primarily to artifacts introduced during amplification. We analyzed the distribution of unique guide sequences relative to the number of reads obtained in the sequencing data, and found that 90.6% of guides were within 1 standard deviation of the mean abundance and 96.1% of guides were within 2 standard deviations, indicating that only a small proportion of the library content is composed of overrepresented sequences (Figure 4C). The guide library is predicted to target 3984 of the 4503 genes annotated in the E. coli genome (88%), grouped by GO Term in Figure 4D. Of the 519 untargeted genes, most are those under 600 nt in length (Figure 4E). Specificity scoring of all guides indicated that 95.3% of guides predicted to arise from this method have a score of 100/100 (Hsu et al., 2013) indicating that only a single location in the E. coli genome is targeted (Figure 4F).
Discussion
In summary, we have outlined a simple method to label chromosomal loci in living samples without altering the genome, and an approach to CRISPR library generation that can be used to produce probes to track any locus or make complex libraries for other purposes. We term this innovative approach to generating guide libraries “CRISPR EATING” (for Everything Available Turned Into New Guides) and anticipate the widespread use of complex guide libraries made using it in a many, perhaps yet unforeseen, applications.
While CRISPR screening libraries generated using synthetic oligonucleotides have been described (Gilbert et al., 2014; Koike-Yusa et al., 2014; Shalem et al., 2014; Wang et al., 2014), so far these libraries target only human and mouse genes. We anticipate that the enzymatic library approach will enable CRISPR-based whole-genome screening in many organisms where oligonucleotide-based design of pooled libraries is undesirable or infeasible for reasons of cost or availability of sequence information. One limitation of our approach is that the precise composition of a guide library cannot be defined as explicitly as it could be in a synthetic oligonucleotide-based library, raising the possibility that individual guides within the library may target more than one location in the genome. We have shown that this is of minimal concern in the small E. coli genome (Figure 4F). For organisms with larger or unsequenced genomes, the use of a cDNA library rather than total genomic DNA as input is likely to decrease the number of low-specificity guides. Furthermore, most screening strategies can tolerate guides that potentially cut at more than one genomic locus because identification of a “hit” mutation is still possible even if several candidate genomic target sites for an isolated guide must be sequenced.
Applying this technique in an imaging context for use in intact cells or embryos represents a practical way to monitor chromosome dynamics in vivo, something that has been an unreachable goal for many years. As the sophistication of libraries generated advances, it may even be possible to monitor whole chromosomes in live samples. One potential concern is that labeling could interfere with normal chromatin compaction. We note that the 3.4 MB region on X. laevis chromosome 4 (Figure 3) is visible with a mean labeling density of one guide per 2,664 bp, which is unlikely to affect global chromatin state. However, because an R-loop (an RNA-DNA hybrid opposite a region of single-stranded DNA) is produced by guide RNA binding, it is possible that nucleosome assembly and chromatin structure are affected (Costantino and Koshland, 2015), particularly in the case of the RHM2 probe that densely labels abundant pericentric repeats.
Methods
Protein purification and designed guide RNA production
dCas9-Neon was expressed as a 230 kD 6xHis-MBP-TEV-dCas9-Neon-Myc fusion protein in BL21 (DE3) Rosetta2 E. coli and affinity purified using Ni-NTA resin, via the Nterminal His tag. The 6xHis-MBP portion of the protein was removed by specific proteolysis using TEV protease to yield the 186 kD dCas9-Neon-Myc.
Xenopus repetitive sequences were scanned for potential dCas9 targeting sites, which included a 5' GG motif for T7 in vitro transcription (IVT) followed by 18–20 nucleotides (nt) of target sequence and a 3' NGG/NAG protospacer motif for CRISPR/Cas9 binding (Cong et al., 2013; Hsu et al., 2013) using Unipro UGENE software (Okonechnikov et al., 2012). This strategy was previously employed in generating sgRNAs for use by injection into zebrafish embryos (Hwang et al., 2009). sgRNAs were synthesized from DNA templates generated by annealing of a ~59 nt 5’ primer containing a T7 RNA polymerase promoter and the desired targeting sequence to an 82 nt 3’ primer containing the necessary invariant sgRNA sequence(Hsu et al., 2013). The 5’ and 3’ primers were annealed over 23 base pairs of reverse-complementarity and extended using a high-fidelity polymerase, resulting in a ~118 base pair double-stranded substrate for use in in vitro transcription reactions. Resulting 100–102 nt RNAs were folded at 60°C and combined with dCas9-Neon at 37°C using 2 µl ~5 mg/ml dCas9 with 5µl IVT reaction product which generally ensured a large molar excess of RNA such that all protein was RNA-bound.
Xenopus egg extract reactions
Cytostatic factor-arrested (CSF) cytoplasmic extracts were prepared from freshly laid eggs of X. laevis and used for spindle assembly reactions as described (Hannak and Heald, 2006). Progression through interphase was induced by addition of 0.5 mM CaCl2 and incubation for 1–2 hours at room temperature. To induce mitotic structures around replicated chromatin, an equal volume of CSF egg extract was then added. High speed metaphase-arrested extracts in which sperm chromatid condensation and resolution occurs were prepared from CSF extracts by centrifugation at 200,000 × g as described (Maresca and Heald, 2006).
Live imaging
Flow cells were prepared using clean microscope slides, double-sided sticky tape (Scott) and coverslips which have been cleaned by sonication for 20 min in ddH2O with detergent (Versa), rinsed and sonicated in ddH2O for 20 min and stored in 70% Ethanol until use (Stehbens et al., 2012). 8–10 µl of extract were used per flow cell. Flow cells were sealed with VaLaP (Vaseline/Lanolin/Paraffin 1:1:1). CSF flow cells were prepared at room temperature, high-speed flow cells on ice just prior to imaging. Extracts were observed through a 60x 1.49 NA Nikon Apochromat oil immersion objective on a customized spinning disk confocal microscope, equipped with a MS-2000 motorized stage (Applied Scientific Instrumentation), a Borealis-modified Yokogawa CSU-X1 spinning disk head (Spectral Applied Research), an LMM5 laser merge module (Spectral Applied Research), automated emission filter changer (Sutter Instrument) and environmental control (In Vivo Scientific). This setup has been described in detail previously (Stehbens et al., 2012). Images were acquired on a iXon low-light electron multiplication CCD (EMCCD) camera at exposure times of 20–50 ms with EM gain set to 150–200 and 3 MHz readout mode. Neutral density filters reducing laser power to 25–50% were used throughout imaging. Microscope and camera were controlled by Nikon Elements Software (Nikon) running on a 64-bit Microsoft Windows 7 PC. Images were analyzed using Fiji (Schindelin et al., 2012) and assembled in Illustrator (Adobe). Pearson’s correlation coefficient was determined using the Coloc2 plugin in Fiji on Z-projections of confocal image stacks; the Pearson’s R value (above threshold) is reported.
sgRNA library construction
The S. pyogenes Cas9 protospacer-adjacent motif (PAM) consists of an NRG motif, where N is any nucleotide, R is an adenine or guanine nucleotide and G is a guanine nucleotide only. To generate DNA ends that are adjacent to this PAM motif, we employed a restriction enzyme cocktail that recognizes a subset of the possible PAMs within a DNA sequence. HpaII, ScrFI and BfaI recognize the sequences C/CGG, CC/NGG and C/TAG respectively, where “/” indicates the site of phosphodiester backbone cleavage. When a substrate is digested with these enzymes and single-strand overhangs are removed, the remaining dsDNA is that existing immediately 5’ to a CGG, NGG or TAG sequence in the target DNA. To trim these blunt-ended PAM-adjacent substrate fragments to 20 nt, we ligated to them an 82 nt dsDNA adapter containing an MmeI recognition site at each terminus, two internal BsaXI sites and an ScrFI site in the middle of the adapter. Following a ligation reaction, products that represent tandems of the adapter are converted back into 82 nt fragments by ScrFI digestion, and those ligated successfully to substrate fragments are trimmed to 41 nt. The 82nt fragments are removed by Ampure XP SPRI size-selection. Because the MmeI enzyme cuts 20 nt distant from its binding site at the end of the ligated adapter, desired substrate fragments are trimmed to 20nt by MmeI digestion, producing a 20 nt substrate fragment 5’ to a 41 nt half-adapter. The resulting fragments are asymmetrical with respect to their single-strand overhangs, with a 2 nt overhang produced by MmeI digestion on one end and a 1 nt overhang produced by ScrFI digestion on the other end. This allows specific ligation of a T7 RNA polymerase promoter to the end produced by MmeI digestion. The T7 RNA polymerase promoter is constructed from two annealed oligonucleotides, one of which has a two nucleotide “NN” (random base) overhang. Following this ligation step, desired fragments now have a T7 promoter, 20 nt of a PAMadjacent region, and 41 nt of an adapter fragment. To produce the final sgRNA fragment, the adapter portion is removed using BsaXI within the adapter. Because BsaXI cuts outside of its recognition site, the position of the BsaXI site permits complete removal of the adapter portion of guide fragments, leaving only a 3 nt overhang. This overhang is exploited for ligation of a 93 nt fragment containing the sgRNA constant region. The resulting 136nt fragment thus consists of a T7 RNA polymerase promoter, 20 nt of sequence corresponding to a putative Cas9 targeting site in the substrate DNA sequence, and 93 nt of sgRNA hairpin. To remove unwanted side products of ligation reactions, the 136 nt fragments were amplified by 10 cycles of PCR using primers in the T7 promoter and at the 3’ end of the sgRNA hairpin. The resulting 136 nt band is isolated and purified using DNA-PAGE, whereupon a second round of 10 cycles of PCR amplification is employed to make the final library.
Computational selection of guides across 3.4MB region on X. laevis chromosome 4
PCR products used to generate the 3.4MB region labeling library used in Figure 3 were selected using a custom computational pipeline employing BioPython to simulate substrate digestion (Cock et al., 2009), BLAST a and a previously published CRISPR scoring algorithm to determine high-scoring guides (Altschul et al., 1990; Camacho et al., 2009; Hsu et al., 2013) and Primer 3 (Untergasser et al., 2012) to generate PCR primers that amplify across regions predicted to produce only high-scoring guides (score of ≥95). Full source code is available at http://github.com/eatingcrispr/ and available in Supplementary Data S1, S2 and S3.
In brief, Scaffold102974 (approx. 21MB) of X. laevis genome v 7.1 was subjected to this computational pipeline. From within the Scaffold, a 3.4MB window containing the largest number of highly specific guides (score of 95+) was used. Within that 3.4MB region, 144 regions containing only guides meeting this score threshold were selected. PCR primers were designed across these regions. A preparation of X. laevis male liver DNA was used as template, and PCRs were carried out using 2X Q5 HotStart Master Mix (New England Biolabs). PCRs were pooled and subjected to the digestion/ligation library protocol using an extended sgRNA constant region.
Supplementary Material
Highlights.
dCas9-Neon is programmed to label repetitive chromosomal loci in egg extracts
Enzymatic processing of any DNA source can generate a guide RNA library
A library generated from PCR products labels a single 3.4 megabase locus
A complex guide RNA library targets the E. coli genome at high frequency
Acknowledgments
We thank the Doudna lab (UC Berkeley) for providing us with a dCas9 cDNA as well as advice on bacterial expression and purification, and Adam Session and Dan Rokhsar for sequence information regarding X. laevis repeats. This work was supported by NIH R01GM098766 (RH) and NIH S10RR26758 (TW). AWG was supported by the National Science Foundation Graduate Research Fellowship Program. A patent is in preparation on CRISPR-EATING (RH, AL). mNeonGreen DNA sequence is licensed under MTA from Allele Biotechnology and Pharmaceuticals to the Regents of the University of California.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: Carried out the experiments: AL, MS, AE, AG. Wrote the paper: AL, RH. Prepared figures: AL, RH, TW, MS, AE.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bassham S, Beam A, Shampay J. Telomere variation in Xenopus laevis. Mol. Cell. Biol. 1998;18:269–275. doi: 10.1128/mcb.18.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST +: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized {CRISPR/Cas} System. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using {CRISPR/Cas} systems. Science (80-.) 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino L, Koshland D. The Yin and Yang of R-loop biology. Curr. Opin. Cell Biol. 2015;34:39–45. doi: 10.1016/j.ceb.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JL, Rayburn AL. Localization of repetitive {DNA} sequences on in vitro Xenopus laevis chromosomes by primed in situ labeling ({PRINS}) J. Hered. 2005;96:603–606. doi: 10.1093/jhered/esi095. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. {CRISPR-mediated} modular {RNA-guided} regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. {Genome-Scale} {CRISPR-Mediated} Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E, Heald R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat. Protoc. 2006;1:2305–2314. doi: 10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. {DNA} targeting specificity of {RNA-guided} Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel S, Meyerhof W, Korge E, Knöchel W. Characterization of highly and moderately repetitive 500 bp Eco {RI} fragments from Xenopus laevis {DNA} Nucleic Acids Res. 1984;12:4921–4938. doi: 10.1093/nar/12.12.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E, Lee J, Jeong J, Park J, Yang Y, Lim J, Kim J, Baek S, Kim K. {SUMOylation} of RORα potentiates transcriptional activation function. Biochem. Biophys. Res. Commun. 2009;378:513–517. doi: 10.1016/j.bbrc.2008.11.072. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable {dual-RNA-guided} {DNA} endonuclease in adaptive bacterial immunity. Science (80-.) 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. {RNA-programmed} genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral {CRISPR-guide} {RNA} library. Nat. Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Maresca TJ, Heald R. Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol. Biol. 2006;322:459–474. doi: 10.1007/978-1-59745-000-3_33. [DOI] [PubMed] [Google Scholar]
- Meyerhof W, Tappeser B, Korge E, Knöchel W. Satellite {DNA} from Xenopus laevis: comparative analysis of 745 and 1037 base pair Hind {III} tandem repeats. Nucleic Acids Res. 1983;11:6997–7009. doi: 10.1093/nar/11.20.6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M, Team U. Unipro {UGENE}: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing {CRISPR} as an {RNA-guided} platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale {CRISPR-Cas9} knockout screening in human cells. Science (80-.) 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens S, Pemble H, Murrow L, Wittmann T. Imaging intracellular protein dynamics by spinning disk confocal microscopy. Methods Enzym. 2012;504:293–313. doi: 10.1016/B978-0-12-391857-4.00015-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the {CRISPR-Cas9} system. Science (80-.) 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. {RNA-guided} genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.