SUMMARY
The genetic programs that maintain leukemia stem cell (LSC) self-renewal and oncogenic potential have been well defined, however the comprehensive epigenetic landscape that sustains LSC cellular identity and functionality is less well established. We report that LSCs in MLL-associated leukemia reside in an epigenetic state of relative genome-wide high-level H3K4me3 and low level H3K79me2. LSC differentiation is associated with reversal of these broad epigenetic profiles, with concomitant down-regulation of crucial MLL target genes and the LSC maintenance transcriptional program that is driven by loss of H3K4me3 but not H3K79me2. The H3K4-specific demethylase KDM5B negatively regulates leukemogenesis in murine and human MLL-rearranged AML cells, demonstrating a crucial role for the H3K4 global methylome in determining leukemia stem cell fate.
Keywords: Epigenome, cancer stem cell, MLL, KDM5B, leukemia
INTRODUCTION
A substantial subset of driver mutations in human cancers targets a variety of epigenetic regulatory factors suggesting that perturbations of the epigenome serve important roles in cancer pathogenesis. These factors include proteins that place, remove, or bind covalent modifications within histones or DNA, and multi-protein complexes that regulate nucleosome position. Perturbations of these factors contribute to localized epigenetic alterations that cause aberrant expression of crucial genes that drive oncogenesis. However, there is much less known about the global epigenetic landscape of cancer cells, particularly the epigenomic features that may distinguish cancer stem cells, the pathologically important subpopulation of cells that sustains disease and mediates relapse.
Emerging evidence based on analyses of a variety of malignancies suggests that global epigenetic abnormalities may be a common feature in human cancer cells (Fraga et al., 2005). For example, epigenomic analysis of the DNA methylomes in two molecular subtypes of chronic lymphocytic leukemia (CLL) showed widespread gene-body DNA hypomethylation when compared to the respective normal B-cell subpopulations (Kulis et al., 2012). In a study of AML epigenomes, an epigenetic signature derived from hundreds of genes with widespread alteration of histone 3 lysine 9 trimethylation (H3K9me3) level was associated with patient prognosis and, when combined with established clinical prognostic markers, outperformed prognosis prediction based on clinical parameters alone (Muller-Tidow et al., 2010). Large scale alterations of cancer cell epigenomes characterized by distinctive histone modifications are also commonly seen in other cancer types including prostate cancer (Seligson et al., 2005), non-small cell lung cancer (Barlesi et al., 2007), low-grade bladder cancer (Barbisan et al., 2008), and renal cell carcinoma (Seligson et al., 2009), in addition to AML (Yamazaki et al., 2013). Broad DNA hypomethylation is present in a variety of cancer types (Kulis and Esteller, 2010). However, it is unclear whether these genome-wide epigenetic features, which are typically measured in bulk tumor cell populations, or partially enriched phenotypic subpopulations, are also a feature of rare cancer stem cells or mechanistically related to their maintenance or differentiation. Therefore, the functional epigenomes of cancer stem cells are generally unknown, and their delineation is compromised by low stem cell prevalence.
Previous studies (Krivtsov et al., 2006; Somervaille et al., 2006) have shown that acute myeloid leukemias induced by MLL oncogenes are organized in a cellular hierarchy whose apex is comprised of a minor fraction of phenotypically distinct progenitor-like leukemia stem cells (LSCs) with self-renewal capability and the potential to differentiate into non-self-renewing progeny cells that constitute the vast majority of the leukemia clone. The ability to enrich for LSCs allowed the demonstration that LSC differentiation and loss of self-renewal is associated with differential expression of several thousand genes (Somervaille, et al., 2009). The current studies were conducted to interrogate the epigenetic landscape of LSCs underlying these broad gene expression changes and determine its role in maintaining LSC oncogenic potential.
RESULTS
LSCs are maintained in an H3K4 hyper-methylation and H3K79 hypo-methylation epigenetic state
To initially interrogate the LSC epigenome we employed a retroviral transduction/transplantation model of AML induced by the MLL-AF10 oncogene (Figure S1A-S1D). In this model, AML cells form a well-defined hierarchy in which sub-populations enriched or depleted for LSCs are distinguished by the presence or absence of c-kit expression, respectively (Somervaille and Cleary, 2006). Clonogenic activity in methylcellulose medium, which is a surrogate marker of LSC potential in this model, showed that LSCs comprised approximately one-quarter of the ckit+ sub-population and were 25 fold more prevalent compared to the more differentiated c-kit− cells.
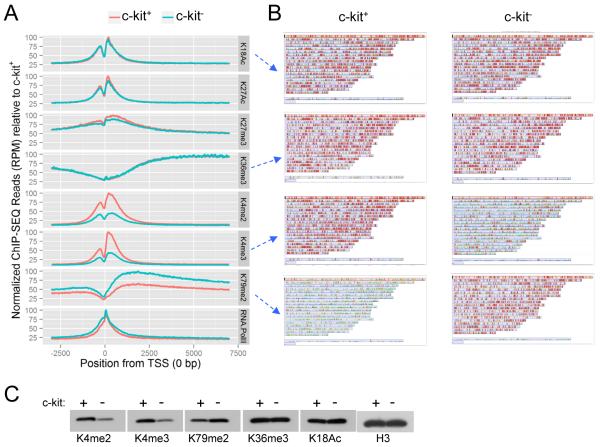
ChIP-seq was performed on the two AML sub-populations using antibodies specific for various histone modifications. Bound DNA regions (ChIP peaks/region) that passed statistical significance were mapped to the genome using a peak-calling algorithm (Table S1). Global read density profiles showed that transcription activation-associated epigenetic marks (H3K4me2, H3K4me3, H3K18ac, and H3K27ac) and the repressive H3K27me3 mark were generally located near the transcription start site (TSS), whereas elongation marks (H3K36me3 and H3K79me2) were predominantly distributed along gene bodies (at significant FDR value, Table S1) (Guenther et al., 2007; Rao et al., 2005).
Assessment of the normalized global ChIP-seq read densities (RPM, reads per million) showed marked differences in the quantitative levels of H3K4 and H3K79 methylation marks in the defined genomic regions (3 kb upstream and 7 kb downstream of TSS) in c-kit+ versus c-kit− cells (Figure 1A). H3K4me2 and H3K4me3 were 60% higher in c-kit+ cells in comparison to c-kit− cells. Conversely, the level of H3K79me2 was approximately 40% lower in c-kit+ cells. All other histone marks were quantitatively similar between the two sub-populations.
Figure 1. Global levels of various histone modifications and RNA Pol II.
(A) Comparison of global levels of various histone marks and RNA Pol II in c-kit+ and c-kit− cells (genomic region −3000 to +7000 relative to TSS). Total number of reads is normalized by RPM (Reads Per Million) for variation between c-kit+ and c-kit− cells. For ease of comparison, RPM is scaled to 100% of c-kit+ for each histone modification or RNA Pol II.
(B) Whole genome heat map view is shown for individual genes with ChIP read density signal encompassing the same genomic region as above.
(C) Western blot analysis was performed on acidic extracted histone proteins of c-kit+ and c-kit− AML subpopulations for the indicated histone modifications.
To interrogate the genomic distribution of the observed differences in histone marks, the ChIP-seq signal in the defined genomic compartment of each individual gene was calculated and plotted as a heat map value on a whole-genome view (Figure 1B and Figure S1E). This showed that H3K4 methylation in c-kit+ cells was distributed broadly throughout the genome and its global reduction in c-kit− cells was not restricted to genes in a specific chromosomal region. H3K79me2 showed an inverse profile with genome-wide quantitative increase from relatively lower levels in c-kit+ to higher in c-kit− cells. All other assessed histone marks (H3K18ac, H3K27ac, H3K36me3, H3K27me3) based on normalized ChIP-seq signals were evenly distributed between c-kit+ and c-kit− cells. Western blot analysis of acid extracted nuclear histones confirmed increased total relative levels of H3K4me2/3 and reduced H3K79me2 in ckit+ versus c-kit− cells (Figure 1C) as also demonstrated by M-A plot analyses (Figure S1F). Thus, c-kit+ cells enriched for LSCs are maintained in a global epigenetic state characterized by relative H3K4 hyper-methylation and H3K79 hypo-methylation. Notably, differentiation of LSCs is associated with inversion of this global epigenetic profile with broad decrease of H3K4me2/3 and increase of H3K79me2.
H3K4 methylation levels exclusively correlate with differential expression of MLL target genes and LSC maintenance genes
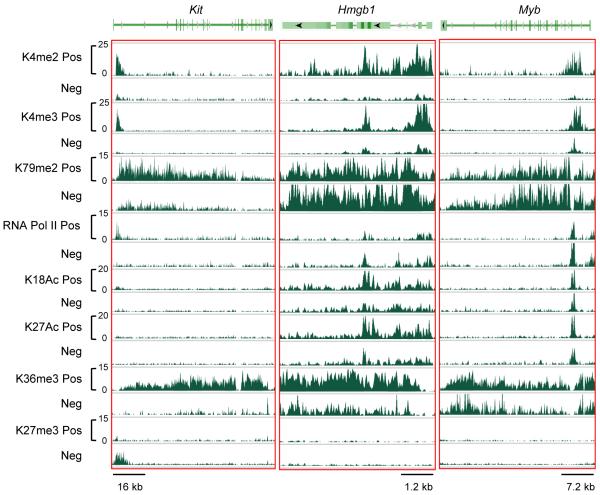
Given the differences in global H3K4 and H3K79 methylation that distinguish c-kit+ cells enriched for LSCs compared to c-kit− cells, we investigated the local histone marks on Hoxa9 and Meis1, which are direct targets of MLL oncoproteins required for AML pathogenesis. Analysis of ChIP-seq read densities showed that Hoxa9 was occupied by activation and elongation histone marks (except H3K36me3), which displayed a broad distribution throughout the adjacent Hoxa gene cluster in both populations (Figure S2). Meis1 was also occupied by active histone marks, including elongation marks, which were distributed throughout the full gene body (Figure S2). The histone marks that most closely correlated with Hoxa and Meis1 expression in the c-kit+ versus c-kit− populations were H3K4me2 and H3K4me3, which were reduced about 40% in the c-kit− LSC-depleted population, consistent with comparably lower transcript levels (Somervaille et al., 2009). H3K79 di-methylation showed anomalous features in both AML subpopulations. It was broadly distributed through the Hoxa and Meis1 genes, and its abundance in c-kit+ cells contrasted with the low global background levels of H3K79me2 but was consistent with that previously observed in bulk MLL leukemia cells (Bernt et al., 2011), likely reflecting aberrant recruitment of the DOT1L histone methyltransferase by MLL-AF10 to its target genes. The c-kit− population unexpectedly showed increased occupancy of H3K79me2 (31% for Hoxa9; 16% for Meis1), which was consistent with increased global background levels, but indicated that reduced Hoxa and Meis1 expression in c-kit− cells was not caused by reduced H3K79 methylation levels.
We also examined the histone marks on genes (Hmgb1 and Myb) previously demonstrated to be differentially expressed in c-kit+ cells and to serve crucial roles in maintaining LSC potential by preventing terminal differentiation (Somervaille et al., 2009). Notably, only H3K4me2/3 marks displayed major variation in density on these genes in the ckit+ versus c-kit− sub-populations (Figure 2). Most other marks demonstrated minimal variation except moderately higher H3K36me3 in c-kit+ cells. The repressive H3K27me3 mark was not present on either gene despite their transcriptional down-regulation. These epigenetic profiles contrasted with those observed on Kit whose differentially expressed product served as a cell surface marker for isolation of LSCs. Kit displayed a more conventional epigenetic profile wherein all active histone marks present in c-kit+ cells were erased in c-kit− cells and replaced with the repressive H3K27me3 mark.
Figure 2. Presence of various histone modifications and RNA Pol II on differentially expressed genes.
Normalized ChIP binding regions are displayed for various histone modifications and RNA Pol II on the Kit, Hmgb1 and Myb genes in c-kit+ and c-kit− cells. The y-axis scale represents normalized read density where c-kit+ and c-kit− cells for individual histone marks use the same scale.
See also Figure S2.
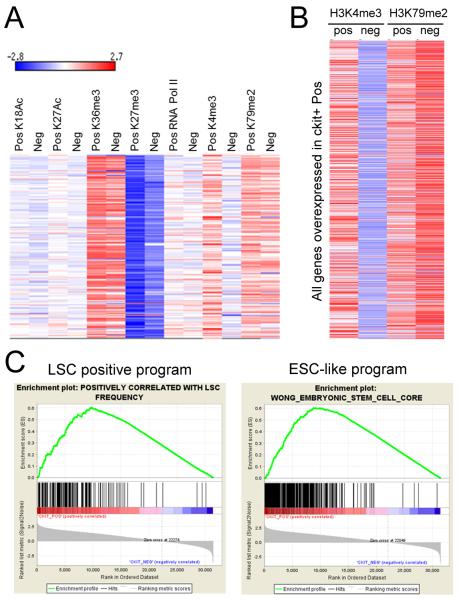
A substantial reduction of H3K4me marks in c-kit− versus c-kit+ cells affected almost all genes of the broader LSC maintenance program (190 genes) (Somervaille et al., 2009) as portrayed in a heat map of mark densities (Figure 3A). Conversely, H3K79me2 levels on these genes were high in c-kit+ cells compared to the low global background levels, and remained relatively high in c-kit− cells suggesting that loss of this mark did not participate in extinguishing the LSC maintenance program to promote LSC terminal differentiation. Assessment of all genes significantly down-regulated in c-kit− versus c-kit+ cells showed a similar H3K4me3 reduction as observed for the LSC maintenance program (Figure 3B) suggesting that global erasure of H3K4me3 was linked to transcriptional down-regulation associated with leukemia cell differentiation.
Figure 3. The LSC positive maintenance transcriptional program is sustained by hypermethylation of H3K4me3.
(A) Heat map displays the densities of histone modification marks and RNA Pol II on genes (190) that comprise the LSC positive maintenance transcriptional program. The ChIP signal density was calculated for full gene length (TSS to gene end) and normalized per total number of reads and per 1 kb of gene length.
(B) Heat map displays the H3K4me3 and H3K79me2 histone modification mark densities on all genes significantly up-regulated in c-kit+ cells as quantified and normalized in panel A.
(C) Gene set enrichment analysis (GSEA) plots showing the LSC positive maintenance program and the ESC-like transcriptional program in genes marked by H3K4me3 in c-kit+ versus c-kit− cells. The data set represents the rank-ordered RefSeq genes with H3K4me3 density ratios in ckit+ versus c-kit− cells.
Gene set enrichment analysis confirmed that genes comprising the LSC maintenance signature were highly enriched for differential H3K4me3 levels in c-kit+ versus c-kit− cells (Figure 3C). Enrichment was also observed for genes encoding an embryonic stem cell-like program driven by MYC and implicated in LSC maintenance. Thus, c-kit+ cells are maintained in a global hyper-methylated H3K4 epigenetic state that sustains the LSC maintenance transcriptional program and the expression of Hoxa and Meis1 genes. Reduced H3K4 methylation is the major epigenetic alteration that correlates with down-regulation of the LSC transcriptional program raising the possibility that global modulation of this mark serves a mechanistic role in LSC maintenance.
Histone demethylase KDM5B negatively regulates leukemia stem cell oncogenic potential
The dynamic changes of the epigenomes in the two AML subpopulations raised the possibility that differential expression of specific writers or erasers of H3K4 methylation marks may regulate LSC potential. Microarray data from published studies (Somervaille et al., 2009) showed that transcript levels of the MLL/SET1 family of HMTs that catalyze H3K4 methylation were not relatively higher in the LSC-enriched subpopulations (Figure S3A). However, transcript levels for Kdm5b, which encodes a member of the KDM5 family of H3K4 histone demethylases, were several-fold higher in c-kit− compared with c-kit+ AML cells (Figure S3B, C). Kdm5b transcripts were also substantially higher in normal differentiated neutrophils compared with bone marrow cells (Figure S3D). These findings raised the possibility that programmed upregulation of KDM5B may extinguish LSC potential by reducing H3K4me2/3 levels on Hox/Meis and LSC maintenance genes.
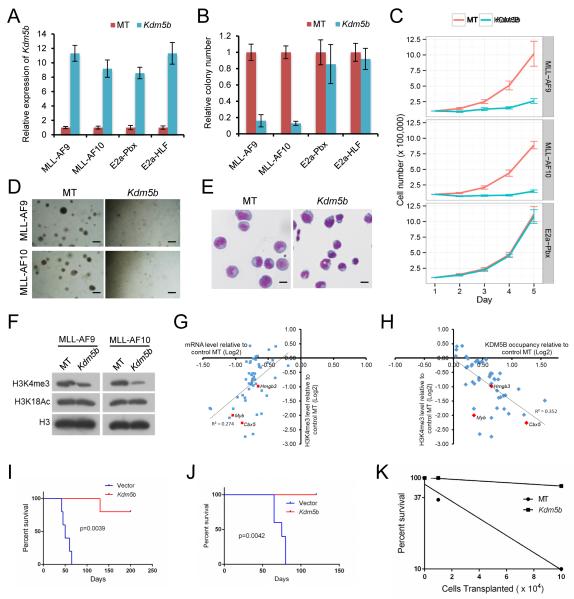
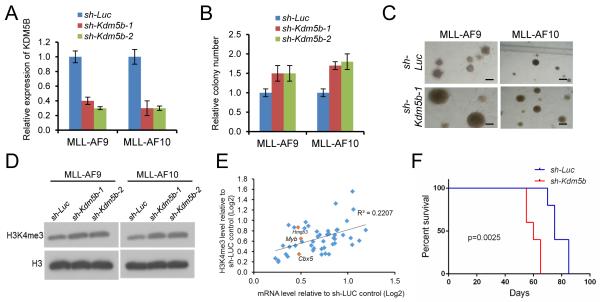
In support of this possibility, ectopic over-expression of KDM5B (Figure 4A) reduced Hoxa9 and Meis1 transcript levels (Figure S3E), and markedly suppressed the in vitro growth of MLL-AF9 transduced cells and MLL-AF10 leukemia cells (Figure 4B-D). Compared with control cells, colony numbers were substantially reduced, and colonies appeared less compact and smaller in size. Cells transformed by non-MLL oncogenes were not significantly affected by forced KDM5B expression, suggesting that the negative effect of KDM5B may be relatively specific to MLL leukemia cells. KDM5B substantially impaired proliferation in liquid culture (Figure 4C), and induced morphological differentiation (Figure 4E). The suppressive effects of KDM5B on CFC potential and H3K4me3 levels were dependent on its catalytic activity (Figure S3F, G). Western blot analysis confirmed that KDM5B forced expression reduced global levels of H3K4me3 in MLL transformed and control cells (Figures 4F and S3H, I). ChIP analysis of MLL-AF10 AML cells demonstrated that most of the top 46 differentially expressed genes comprising the LSC maintenance program displayed reduced H3K4me3 levels and increased KDM5B occupancy that generally correlated with reduced transcript levels in response to KDM5B forced expression (Figure 4G, H). Transplantation of KDM5B over-expressing cells into syngeneic recipient mice showed a marked reduction in leukemia penetrance and increased survival compared with vector control cells (Figure 4I, J). Limit-dilution secondary transplantation assays showed that the LSC frequency in MLL-AF10 AML was reduced more than 15-fold in response to KDM5B over-expression (Figure 4K). Thus, KDM5B suppresses leukemogenesis and forces AML cells to differentiate out of the LSC compartment.
Figure 4. KDM5B reduces global H3K4me2/3 methylation and suppresses AML.
(A) Bar graph shows Kdm5b transcript levels in transformed cells (denoted below) transduced with the indicated KDM5B over-expression or control MT (empty) vectors. Transcript levels were quantified by RT-PCR at 2 days and expressed relative to control vector.
(B, C) Growth in methylcellulose (B) or liquid (C) culture was quantified after exogenous over-expression of KDM5B in cells transformed by the indicated oncogenes. Colony counts are expressed relative to empty vector (MT) transduced control cells.
(D) Representative colonies are shown for experiment in panel B. Scale bar represents 20 μm.
(E) Light microscopy of May-Grunwald/Giemsa-stained leukemia cells after transduction with KDM5B over-expression constructs. Scale bar represents 10 μm.
(F) Western blot analysis was performed using the antibodies indicated on the left to assess global histone modification levels following exogenous over-expression of KDM5B and empty vector (MT).
(G) Scatter plot is shown for H3K4me3 and mRNA expression levels on 46 genes in the LSC program of MLL-AF10 AML cells transduced with control empty vector (MT) or KDM5B over-expression constructs. Correlation is indicated as R2 value.
(H) Scatter plot is shown for H3K4me3 and KDM5B occupancy levels on 46 genes in the LSC program of MLL-AF10 AML cells transduced with control (MT) or KDM5B over-expression constructs. Correlation is indicated as R2 value.
(I, J) Survival curves are shown for cohorts of mice transplanted with MLL-AF9 transformed cells (I) or MLL-AF10 AML cells (J) transduced with control or KDM5B over-expression constructs (n = 5 each cohort). Acute leukemia was confirmed by peripheral blood leukocyte count or necropsy. Log-rank Test was used for statistical analysis.
(K) Limit-dilution transplantation analyses performed on MLL-AF10 leukemia cells transduced with control (MT) or KDM5B over-expression constructs indicate cell numbers required to initiate AML in sublethally irradiated recipient mice (n=3 for each cell dose).
Error bars represent SEM.
See also Figure S3.
Knockdown of KDM5B had opposite effects on MLL-transformed cells. In methylcellulose assays, reduced levels of KDM5B (Figure 5A) caused a significant increase in colonies that were substantially larger and more densely compact, indicating that they contained increased numbers of progenitors impaired in their differentiation (Figure 5B, C). KDM5B knockdown was associated with increased levels of H3K4me3, both globally (Figure 5D) and more specifically on the top 46 genes of the LSC program, which showed concomitant increases in their transcript levels (Figure 5E). A substantial enhancement in leukemia aggressiveness was observed following transplantation of KDM5B knockdown MLL-AF10 cells as evidenced by a significantly decreased latency for AML development (Figure 5F). These data support a tumor suppressor role for KDM5B, and modulation of its expression to perturb the epigenome in MLL leukemias demonstrates the importance of H3K4 methylation levels in maintaining LSC potential.
Figure 5. Knockdown of KDM5B promotes AML and LSC potential.
(A) Bar graph indicates Kdm5b transcript levels in cells transduced with the indicated shRNAs. Transcript levels were quantified by RT-PCR at 2 days and expressed relative to control vector.
(B) Growth in methylcellulose culture was quantified after shRNA knockdown of KDM5B in cells transformed by the indicated oncogenes. Results are expressed relative to control shRNA transduced cells.
(C) Representative colonies are shown for experiment in panel B. Scale bar represents 50 μm.
(D) Western blot analysis was performed using the antibodies indicated on the left to assess global histone modification levels following knockdown of KDM5B.
(E) Scatter plot is shown for H3K4me3 and mRNA expression levels on 46 genes in the LSC program in MLL-AF10 AML cells transduced with control or Kdm5b shRNA constructs, their correlation is indicated as R2 value.
(F) Survival curves are shown for cohorts of mice transplanted with MLL-AF10 AML cells transduced with control or Kdm5b shRNA constructs (n = 5 each cohort). Acute leukemia was confirmed by peripheral blood leukocyte count or necropsy. Log-rank Test was used for statistical analysis (p = 0.0025).
Error bars represent SEM.
Histone demethylase KDM5B negatively regulates human MLL leukemia cells
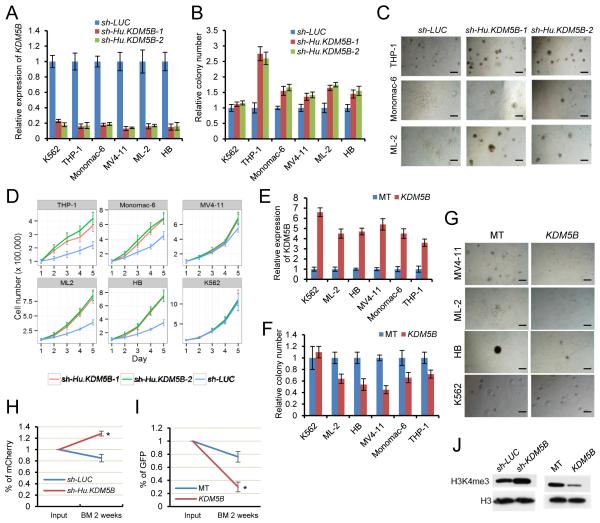
A similar approach was employed to investigate whether the H3K4 epigenome serves an analogous role in sustaining oncogenic potential of human MLL-rearranged (MLLr) leukemias. KDM5B was knocked down in various human leukemia cell lines harboring several different MLL fusion genes including THP-1 (MLL-AF9), Monomac-6 (MLL-AF9), MV4-11 (MLL-AF4), ML-2 (MLL-AF6), and HB (MLL-ENL), as well as the non-MLLr leukemia cell line K562. In MLLr leukemias, an ~80% knockdown of KDM5B induced a significant increase in blast-like colonies that were substantially larger and more densely compact compared with controls, consistent with the presence of increased numbers of progenitors with impaired differentiation (Figure 6A-C). In contrast, K562 cells were not affected (Figure S4A). Knockdown MLLr leukemia cells also proliferated faster (less so for MV4-11) in liquid culture (Figure 6D) further indicating that KDM5B normally serves a leukemia suppressive role. Consistent with this, forced expression of KDM5B (fused with GFP to provide a selectable marker) reduced colony formation in MLLr leukemia cells (Figure 6E, F) and remaining colonies appeared smaller with more differentiated morphologies compared with controls (Figure 6G). Non-MLLr cells (K562, Kasumi-1, and HL-60) did not show reductions in colony numbers following KDM5B over-expression (Figure 6F, 6G, S4B, C). HOXA9 and MEIS1 gene transcripts and their H3K4me3 levels were significantly reduced only in MLLr leukemia cells following KDM5B over-expression despite global reduction in H3K4me3 levels (Figure S4D-F), which partly explains the selective effect of KDM5B in MLLr leukemia cells.
Figure 6. KDM5B regulates the oncogenic potential of human MLL leukemia cells.
(A) Bar graph indicates KDM5B transcript levels in cells transduced with the indicated shRNAs. Transcript levels were quantified by RT-PCR at 2 days and expressed relative to control vector.
(B) Growth in methylcellulose was quantified after shRNA knockdown of KDM5B in the indicated human leukemia cell lines. Colony counts are expressed relative to those of control shRNA.
(C) Representative colonies are shown for experiment in panel B. Scale bar represents 20 μm.
(D) Proliferation in liquid culture was measured after shRNA knockdown of KDM5B in the indicated human leukemia cell lines.
(E) Bar graph indicates KDM5B transcript levels in cells transduced with KDM5B over-expression or control empty (MT) vectors. Transcript levels were quantified by RT-PCR at 2 days and expressed relative to control vector.
(F) Growth in methylcellulose was quantified after exogenous over-expression of KDM5B in the indicated human leukemia cell lines. Colony counts are expressed relative to those of vector control.
(G) Representative colonies are shown for experiment in panel F. Scale bar represents 20 μm.
(H and I) In-vivo engraftment assay shows the relative abundance of MV4-11 cells transduced with KDM5B knockdown (H) or over-expression (I) lentiviral constructs in the bone marrow at two weeks post-transplantation. Results represent the percentage of transduced cells (mCherry or GFP positive) within the total CD45+ population and are expressed relative to the percentage in the input population immediately prior to transplantation (*, p < 0.01)
(J) Western blot analysis shows H3K4me3 levels in flow sorted transduced MV4-11 cells following KDM5B knockdown (mCherry+ cells) or over-expression (GFP+ cells).
Error bars represent SEM.
See also Figure S4.
Xenotransplantation studies showed that knockdown of KDM5B enhanced bone marrow engraftment efficiency of MV4-11 cells in NSG mice by about 40% compared to control (sh-LUC) cells (Figure 6H), whereas over-expression of KDM5B substantially impaired engraftment (Figure 6I) at two weeks. Western blot analysis of FACS-sorted transduced cells prior to transplant confirmed the respective increase and decrease of global H3K4me3 levels in KDM5B knockdown and over-expression cells (Figure 6J).
KDM5B suppresses LSC activity in human MLL-rearranged AML
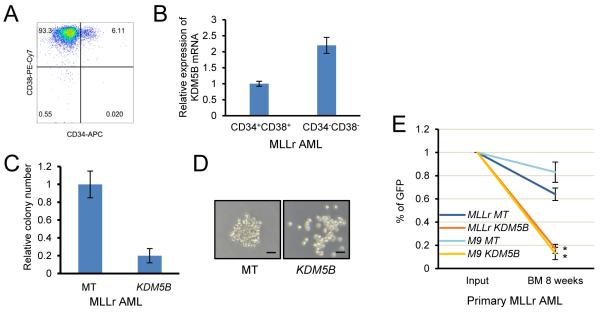
The role of KDM5B was assessed in primary human AML, wherein LSCs are typically enriched in the CD34+ compartment including the CD34+CD38− and CD34+CD38+ subfractions (Eppert et al., 2010). We first compared KDM5B expression in prospectively purified LSC-enriched (CD34+CD38+) and LSC-depleted (CD34−CD38−) MLLr AML subpopulations (Figure 7A), which showed KDM5B upregulation in the more differentiated double negative cells consistent with a tumor suppressive role in human AML (Figure 7B). In support of this, primary MLLr AML cells transduced with a KDM5B over-expression lentivirus formed significantly fewer colonies and their morphology appeared less compact and more differentiated than the blast-type colonies prevalent in the vector control culture (Figure 7C, D). In addition, xenotransplantation studies of two primary MLLr AMLs showed that enforced expression of KDM5B substantially reduced bone marrow engraftment efficiency in NSG mice compared to controls (empty vector) (Figure 7E). These results support that histone demethylase KDM5B negatively regulates the oncogenic potential of human MLLr leukemia cells analogous to its role observed in mouse LSCs.
Figure 7. KDM5B suppresses the oncogenic potential of human MLLr AML.
(A) Flow cytometry analysis shows CD34 and CD38 expression by a representative human MLLr AML.
(B) KDM5B transcript levels were quantified by RT-PCR analysis of flow-sorted MLLr AML cells with the indicated CD34/CD38 phenotypes. Results are expressed relative to levels in the CD34+CD38+ population and represent the mean and SEM of three replicates.
(C) Bar graph indicates growth in methylcellulose culture of MLLr AML cells transduced with KDM5B or lentiviral empty vector (MT) control. Results are expressed relative to empty vector control cells.
(D) Representative colonies are shown for experiment in panel C. Scale bar represents 100 μm.
(E) In-vivo engraftment assay shows the relative abundance of two primary MLLr AMLs transduced with KDM5B over-expression lentiviral constructs in the bone marrow at eight weeks post-transplantation. Results represent the percentage of transduced cells (GFP+) within the total CD45+ population and are expressed relative to the percentage in the input population immediately prior to transplantation (*, p < 0.01).
Error bars represent SEM.
DISCUSSION
Genome-wide epigenetic mechanisms are well known to serve crucial roles in normal stem cell biology. However, a comprehensive analysis of functionally validated cancer stem cell epigenomes has been hindered by limited quantities of sufficiently enriched stem cells to perform similar genome-wide analyses. Thus, it has been unclear if the epigenomes of cancer stem cells are distinct from more differentiated progeny cells in the tumor that lack stem cell activity, nor is it clear whether the biological properties that define malignant stem cells are maintained or extinguished by global features of the epigenome. In this report, we demonstrate that the c-kit+ population of leukemia cells enriched for LSCs (25%) is maintained in a distinctive epigenomic state that undergoes global change with their subsequent differentiation. The epigenomes of c-kit+ cells share features with ESCs and HSCs including a major role for the global state of H3K4 tri-methylation in regulating LSC fate and oncogenic potential. Mechanistically, this is orchestrated by the histone demethylase KDM5B, which functions as a tumor suppressor by down-regulating transcriptional programs that otherwise sustain LSC self-renewal and leukemogenic function.
H3K4me3 is a functionally important histone mark that is typically promoter centric in location, associated with open chromatin, and generally present on actively transcribed genes. Global H3K4 methylation state has been implicated in normal stem cell functions, particularly the maintenance and differentiation of ESCs (Bernstein et al, 2006; Mikkelsen et al, 2007; Meissner et al, 2008). Both mouse and human ESCs display globally more open and dynamic chromatin characterized by enrichment of widespread active chromatin domains containing H3K4me3 (Ho and Crabtree, 2010; Meshorer et al., 2006; Azuara et al., 2006; Lister et al., 2009), and global loss of H3K4me3 accompanies differentiation of ESCs to progenitor cells (Efroni et al., 2008).
In HSCs, about two-third of genes are occupied by H3K4me3 (Sun et al., 2014; Cui et al., 2009) and upon induced differentiation thousands of genes lose this active mark (Cui et al., 2009). Analysis of chromatin marks in isolated hematopoietic cell subpopulations (Lara-Astiaso et al., 2014) shows that high global H3K4me3 undergoes reduction (30-40%) with differentiation of HSCs to granulocytes and monocytes (data not shown). In addition, enhancement of H3K4me3 on important HSC genes by knockdown of KDM5B antagonizes HSC differentiation (Cellot et al., 2013). These findings are consistent with our observations that KDM5B expression is upregulated with normal myeloid differentiation. Similarly, in c-kit+ cells enriched for LSCs several thousand genes are differentially expressed upon differentiation and over one-third of them are down-regulated in parallel with reduced H3K4me3 suggesting that the LSC transcriptional program is epigenetically maintained by H3K4me marks whose broad reduction induces LSCs to differentiate. These studies support a model wherein differentiation of both normal hematopoietic cells and AML cells is accompanied by programmed reduction of global H3K4 methylation. This differentiation-linked epigenetic program antagonizes LSC oncogenic (and HSC) potential, but is opposed by MLL oncoproteins with which it competes. MLLr leukemias are highly sensitive to alterations in the H3K4 methylome since the MLL-specific gene program is inordinately dependent on H3K4 methylation status, particularly key target genes such as HOXA9 and MEIS1 whose expression is normally maintained by WT MLL, an H3K4 methyltransferase that is also required for MLL-mediated transformation (Thiel et al., 2010) likely contributing to the selective role of H3K4me in this genetic subtype of AML compared with non-MLLr AML. Thus, LSC differentiation shares epigenetic features with normal stem cells, wherein broad reduction of H3K4 methylation extinguishes the HSC program and serves as a primary oncogenic off-switch to down-regulate the LSC program in MLLr leukemia cells.
Our studies demonstrate that the histone demethylase KDM5B is a negative regulator of the oncogenic state through its effects on the crucial effector genes that otherwise sustain LSC potential in AML. This is consistent with recent studies that chromatin regulators may modulate transcriptional responses underlying cell fate decisions and cellular differentiation, and in maintaining the identity of differentiated cells (Chen and Dent, 2014). KDM5B specifically demethylates H3K4me1/2/3 (Yamane et al., 2007; Pedersen and Helin, 2010). It is essential for early embryonic development (Catchpole et al, 2011) and important for ESC differentiation. KDM5B depletion has been shown to block differentiation along the neural lineage due to increased global H3K4me3 levels and consequent failure to efficiently silence stem and germ cell related genes (Schmitz et al., 2011). Depletion of KDM5B also impairs ESC differentiation to embryoid bodies causing partial retention in a self-renewing stage (Kidder et al., 2013). These studies are consistent with our observations that KDM5B depletion antagonizes leukemia cell differentiation, upregulates expression of LSC maintenance genes, and substantially enhances the malignant phenotypes of mouse and human leukemia cells. LSCs are maintained in a state of high global H3K4 methylation and low KDM5B expression, whereas upregulation of KDM5B induces leukemia cell differentiation, concomitant loss of LSC fate, and down-regulation of leukemia maintenance genes and key MLL target genes.
The function of KDM5B as a suppressor of MLL leukemia contrasts with its potential role in other cancer types where it is inferred to have oncogenic activity based primarily on elevated levels of expression in several solid cancers including breast (Lu et al., 1999), prostate (Xiang et al., 2007) and bladder (Hayami et al., 2008). Conversely, KDM5B under-expression in an aggressive subtype of breast cancer (Yamane et al., 2007; Qian et al., 2011) and primary melanoma is consistent with a possible tumor suppressive function (Roesch et al., 2008). In melanoma, high level KDM5B expression in a subpopulation of slow-cycling cells that maintain a high proliferative population suggested a potential oncogenic role in cancer stem cells (Roesch et al., 2010). Taken together, the expression profiles of KDM5B suggest that its specific oncogenic contributions may be context dependent. However, our studies raise caution about assigning a functional role for KDM5B or other epigenetic factors based on expression in bulk tumor cell populations where cancer stem cell specific attributes may be obscured. Through analysis of enriched AML subpopulations our studies reveal a dynamic expression profile within the leukemia cell hierarchy wherein high level KDM5B expression in the majority of leukemia cells is not oncogenic, but rather mediates tumor suppressive function promoting differentiation by extinguishing the transcriptional program that otherwise sustains LSC potential.
In addition to a functionally dynamic H3K4 methylome in MLL leukemia, c-kit+ LSC-enriched cells display low levels of global H3K79 methylation, which convert to higher levels in the LSC-depleted subpopulation. H3K79me2 is a crucial epigenetic mark required for leukemogenesis induced by MLL fusion proteins, which recruit DOT1L, the methyltransferase that writes the H3K79me2 mark, and other elongation factors to deregulate expression of key MLL target genes (Okada et al., 2005; Krivtsov et al., 2008). Our finding of high H3K79me2 occupancy on Hoxa9 and Meis1 genes in the context of low background levels is consistent with previous observations in bulk MLL leukemia cells (Bernt et al., 2011), and reinforces the role of its aberrant deposition on MLL target genes in leukemia pathogenesis. The increase in H3K79me2 on MLL target and LSC maintenance genes in the LSC-depleted population is unexpected since it runs counter to their reduced expression and loss of LSC potential during AML cell terminal differentiation. However, the effects of increased H3K79me2, an elongation mark, on target gene expression may be negated by concomitant loss of H3K4me3, an upstream initiation mark. The differences in genome-wide H3K79 methylation are not likely caused by differences in steady state levels of DOT1L, based on comparable transcript levels (data not shown), but a potential role for suppressed demethylation cannot be excluded since a specific H3K79 demethylase has not been identified. Alternatively, global H3K79me2 levels may reflect the relative function and/or abundance of WT AF10 given its role in DOT1L recruitment and stimulation (Deshpande et al., 2014). Although the mechanism underlying increased global levels of H3K79me2 in MLLr c-kit− cells is unclear, relatively high levels of this mark in the LSC-depleted subpopulation correlate with higher global levels of the H3K36me3 elongation mark and Pol II consistent with increased transcription of genes that promote myeloid differentiation. Anti-DOT1L efficacy with small molecular inhibitor has been observed in preclinical studies (Daigle et al, 2011) indicating that the inhibitory effects on MLL target gene expression prevail over potential inhibitory effects on AML cell differentiation. Our studies provide a rationale for an alternative or complementary therapeutic strategy targeting deposition of H3K4me3, which serves a major global epigenetic role in sustaining MLL leukemia and LSC potential.
EXPERIMENTAL PROCEDURES
Cell culture
Human leukemia cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 1× penicillin/streptomycin/L-glutamine (PSG), and 1× non-essential amino acids (NEAA). Transformed mouse myeloid cells or leukemia cells were cultured in RPMI 1640 medium supplemented with 20% FBS, 20% WEHI-conditioned medium, and PSG (R20/20 medium), or in methylcellulose-containing medium (Methocult M3231, Stem Cell Technologies) with cytokines as previously described (Lavau et al., 1997). HEK-293T and Phoenix-Eco cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FBS and PSG.
Isolation of leukemia cells for ChIP-seq
Leukemias were induced by retroviral transduction/transplantation of c-kit+ BM stem and progenitor cells with MLL-AF10 as previously described (Somervaille and Cleary, 2006). For ChIP-seq, leukemia cells were collected from spleens and immediately fractionated based on ckit expression using CD117 microbeads (130-091-224, Miltenyi Biotec) in an autoMACS according to the manufacturer’s protocol. Enrichment of CFCs was quantified by plating in methylcellulose-containing medium with cytokines (Somervaille and Cleary, 2006). Fractionated cells were fixed at room temperature for 10 minutes with 0.7 % formaldehyde solution (F8775, Sigma-Aldrich) in R20/20 medium and frozen at −80°C until used for ChIP. All experiments on mice in this study were performed with the approval of, and in accordance with, the Stanford University Administrative Panel on Laboratory Animal Care.
ChIP-seq and bioinformatics analyses
Fixed leukemia cells were lysed for 20 minutes in nuclear lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS) containing freshly added 1× proteinase inhibitor cocktail (Roche) on ice, then sonicated to generate DNA fragments of 150-500 base pair using a Diagenode Bioruptor XL sonicator for a total duration of 60 cycles (30 seconds ON and 30 seconds OFF). The soluble chromatin was diluted 10-fold with ChIP dilution buffer (20m M Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.01% SDS, 1% Triton-X 100) containing 1× proteinase inhibitor cocktail, and the diluted chromatin (1 mL equivalent to 2 million cells) was subjected to immunoprecipitation using various ChIP grade antibodies coupled with protein A magnetic beads (Invitrogen). After overnight immunoprecipitation at 4°C, washes and reverse crosslinking as well as ethanol precipitation of ChIP DNA were performed by standard ChIP assay protocol. DNA was used for ChIP-seq library construction based on manufacturer’s protocol with modifications to account for limited number of cells (Schmidt et al., 2009). Sequencing was carried out at the Stanford Genome Center.
The sequencing reads were mapped to the mouse genome and statistically confident mapped reads were used for ChIP peak/region calling using Quest (Valouev et al., 2008) and DNANexus (https://www.dnanexus.com), which also provided genome browsing. Downstream analysis including genome-wide localization density plot, chromosome-view of global ChIP signal heat map, and M-A Plot were performed by using a combination of tools including DNANexus, Seqmonk (http://www.bioinformatics.babraham.ac.uk/), and R program (R Development Core Team (2012). Some graphs were plotted using the R program package, ggplot2 (Wickham 2009). Cluster analysis of H3K4me3 and H3K27me3 was performed using seqMINER (Ye et al., 2011). GO term analysis of bivalent genes was performed using DAVID Functional Annotation Database (Huang et al., 2009). For gene set enrichment analysis (GSEA) of ChIP-seq data, the ranked genome-wide genes based on their ChIP-seq signal calculated by normalized ChIP-seq reads on full length genes were treated as a data set for performing enrichment analysis of individual gene sets using 1,000 data permutations.
For standard ChIP assays, chromatin equivalent to 5 million fixed cells was used for immunoprecipitation. Primers used for target gene detection are provided in Supplemental Experimental Procedures.
Leukemogenesis assays
For over-expression studies, mouse MLL-AF10 leukemia cells or MLL-AF9 immortalized cells were transduced with MSCV- puro retrovirus expressing full length mouse KDM5B or empty vector. Transduced cells were cultured in R20/20 medium for 48 hours, then selected in puromycin for 24 hours, prior to transplantation (106 cells) into sub-lethally irradiated (450 rad) C57BL/6 mice. For limit dilution assays, serial dilutions (105, 104, 0) of MLL-AF10 leukemia cells transduced with MSCV-KDM5B-puro or empty vector were transplanted into sub-lethally irradiated mice. For knockdown studies, mouse MLL-AF10 leukemia cells transduced with pSicoR-mCherry lentivirus expressing shRNAs were cultured 2-3 days in R20/20 before FACS isolation based on mCherry fluorescence. The sorted mCherry positive cells were cultured for several passages before transplantation (1.5×106 cells) into sub-lethally irradiated recipients.
Viral vectors and transduction
shRNAs were designed using a commercial web tool (Invitrogen) and consisted of the following: sh-mKDM5B-1 (GCAGTTGTTTGCAAGGATAGA), sh-mKDM5B-2 (GGGAGTAATTGATTCTGAAAG), sh-hKDM5B-1 (GGAGATGCACTTCGATATA), shhKDM5B-2 (GCTTCCAAGGCTGATGTATTA). The shRNAs were cloned into pSicoR-mCherry lentivirus vector. Lentiviruses were generated by co-transfection of shRNA constructs into HEK-293T cells with pCMV-dR8.2 (packaging) and pCMV-VSVG (envelope) plasmids. Mouse and human cells were transduced by spinoculation and cultured for 2-3 days prior to FACS isolation based on mCherry fluorescence. Mouse full length Kdm5b cDNA was subcloned into MSCV-puro from pUB6-V5-KDM5B, which was kindly provided by Dr. Impey (Xie et al., 2011). Retroviral plasmids were packaged in Phoenix-Eco cells. Viral supernatants were concentrated (at least 10-fold) using retro-x concentrator (Clontech), and supplemented with polybrene for transduction of mouse leukemia cells by spinoculation (2500 rpm, 32°C for 2.5 hour).
Xenotransplantation studies
MV4-11 cells were transduced with pSicoR-mCherry encoding KDM5B shRNAs or GFP-tagged KDM5B cDNA. The percentage of fluorescent cells was determined at 48 hours by flow cytometry, and three million total cells were then transplanted into sublethally irradiated (200 rad) NSG mice. Two weeks later, bone marrow cells were collected and analyzed by flow cytometry for the percentage of engrafted human cells (CD45+) expressing the respective fluorescent marker. Primary human MLLr AML cells (cryopreserved in the Pediatric Cell Bank at Stanford) were obtained with informed consent and institutional review board approval. Cryopreserved MLL-rearranged primary AML samples were thawed and cultured in IMDM with 15% FBS supplemented with cytokines (10 ng/ml hSCF, 10 ng/ml hFlt3, 10 ng/ml hIL-7, 10 ng/ml hIL-3, 10 ng/ml hIL-6, 5 ng/ml SR1) for two hours. Cells were then transduced with lentivirus expressing GFP-tagged KDM5B. The percentage of fluorescent cells was determined 48 hours after transduction, and one million cells were immediately transplanted into sublethally irradiated (200 rad) NSG mice. Eight weeks later, bone marrow cells were collected and analyzed by flow cytometry for the percentage of engrafted human cells (CD45+) expressing GFP.
Real-time PCR
RNA was isolated using TRIzol reagent (15596-026, Invitrogen) and converted to cDNA using SuperScript III First-Strand Synthesis System (Life Technologies). Quantitative PCR analysis was performed using an ABI StepOnePlus real time PCR machine. Primers used for SYBR detection are listed in Supplemental Experimental Procedures. All signals were quantified using the ΔCt method and were normalized to the levels of GAPDH (Gapdh) or Beta-actin.
Western blot analysis
Western blots for histone modifications were performed on acid extracted nuclear proteins. The following antibodies used for western blot and ChIP assays were obtained from Abcam: anti-H3K4me2 (ab7766); anti-H3K4me3 (ab8580); anti-H3K18ac (ab1191); anti-H3K27ac (ab4729); anti-H3K36me3 (ab9050); anti-H3K79me2 (ab3594); and anti-RNA Pol II (ab817). Anti-H3K27me3 (07-449) was obtained from Millipore.
Supplementary Material
SIGNIFICANCE.
Localized epigenetic alterations serve crucial roles in causing aberrant regulation of primary target genes for MLL and other oncoproteins, however much less is known about the global epigenetic landscape of leukemia cells, particularly the epigenomic features that may functionally distinguish leukemia stem cells (LSCs), the pathologically important subpopulation of leukemia cells that sustains disease and mediates relapse. In this report the authors demonstrate that the global state of H3K4 tri-methylation serves a major role in regulating LSC fate and oncogenic potential. Mechanistically, this is orchestrated by the histone demethylase KDM5B, which functions as a tumor suppressor to extinguish the LSC maintenance transcriptional program.
ACKNOWLEDGMENTS
We thank Maria Ambrus and Cita Nicolas for technical assistance, and members of the Cleary lab for helpful discussions. We thank Anton Valouev for advice on ChIP-seq peak calling, and Kathy Sakamoto and Norman Lacayo for providing primary human MLLr cells. S. H. K. W. was supported by a Yong Investigator Award from the Alex’s Lemonade Stand Foundation for Childhood Cancer. H.-P.K. was supported by PHS Grants T32-CA09302 and T32-CA09151, awarded by the National Cancer Institute, DHHS and Dean’s Postdoctoral Fellowship at the Stanford School of Medicine. J. D.-A. was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft, ref. DU 1287/2-1). We acknowledge support from the Children’s Health Initiative of the Packard Foundation, and Public Health Service (PHS) grant CA116606.
Footnotes
ACCESSION NUMBERS
The Gene Expression Omnibus (http://ncbi.nlm.nih.gov/geo) accession number for the ChIP-seq data reported in this article is GSE60193).
Supplemental Information includes four figures and three tables.
AUTHOR CONTRIBUTIONS
S.H.K.W. and M.L.C. developed the hypothesis and wrote the manuscript. S.H.K.W. designed and performed experiments, analyzed data, and conducted bioinformatics analyses. D.L.G. advised on data analysis. M.I., M.C.W., D.S., H-P.K., L.Z. and Z.W. performed and interpreted experiments. J.D-A. reviewed and discussed results, and contributed to manuscript preparation. M.L.C. supervised the project.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nature Cell Biology. 2006;8:532–U189. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Barbisan F, Mazzucchelli R, Santinelli A, Stramazzotti D, Scarpelli M, Lopez-Beltran A, Cheng L, Montironi R. Immunohistochemical evaluation of global DNA methylation and acetylation in papillary urothelial neoplasm of low malignant potential. International Journal of Immunopathology and Pharmacology. 2008;21:615–623. doi: 10.1177/039463200802100315. [DOI] [PubMed] [Google Scholar]
- Barlesi F, Giaccone G, Gallegos-Ruiz MI, Loundou A, Span SW, Lefesvre P, Kruyt FAE, Rodriguez JA. Global histone modifications predict prognosis of resected non-small-cell lung cancer. Journal of Clinical Oncology. 2007;25:4358–4364. doi: 10.1200/JCO.2007.11.2599. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie XH, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng ZH, Punt N, Daigle A, Bullinger L, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchpole S, Spencer-Dene B, Hall D, Santangelo S, Rosewell I, Guenatri M, Beatson R, Scibetta AG, Burchell JM, Taylor-Papadimitriou J. PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. International Journal of Oncology. 2011;38:1267–1277. doi: 10.3892/ijo.2011.956. [DOI] [PubMed] [Google Scholar]
- Cellot S, Hope KJ, Chagraoui J, Sauvageau M, Deneault E, MacRae T, Mayotte N, Wilhelm BT, Landry JR, Ting SB, et al. RNAi screen identifies Jarid1b as a major regulator of mouse HSC activity. Blood. 2013;122:1545–1555. doi: 10.1182/blood-2013-04-496281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TP, Dent SYR. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nature Reviews Genetics. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui KR, Zang CZ, Roh TY, Schones DE, Childs RW, Peng WQ, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AJ, Deshpande A, Sinha AU, Chen L, Chang J, Cihan A, Fazio M, Chen CW, Zhu N, Koche R, et al. AF10 regulates progressive H3K79 methylation and HOX gene expression in diverse AML subtypes. Cancer Cell. 2014;26:896–908. doi: 10.1016/j.ccell.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RDG, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, Metzeler KH, Poeppl A, Ling V, Beyene J, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nature Medicine. 2011;17:1086–U1091. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature Genetics. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayami S, Yoshimatsu M, Veerakumarasivam A, Unoki M, Iwai Y, Tsunoda T, Field HI, Kelly JD, Neal DE, Yamaue H, et al. Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Molecular Cancer. 2010;9 doi: 10.1186/1476-4598-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kidder BL, Hu GQ, Yu ZX, Liu CY, Zhao KJ. Extended Self-Renewal and Accelerated Reprogramming in the Absence of Kdm5b. Molecular and Cellular Biology. 2013;33:4793–4810. doi: 10.1128/MCB.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Kulis M, Esteller M. DNA Methylation and Cancer. In: Herceg Z, Ushijima T, editors. Epigenetics and Cancer, Pt A. 2010. pp. 27–56. [DOI] [PubMed] [Google Scholar]
- Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G, Martinez-Trillos A, Castellano G, Brun-Heath I, Pinyol M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nature Genetics. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, et al. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. Embo Journal. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q-M, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PJ, Sundquist K, Baeckstrom D, Poulsom R, Hanby A, Meier-Ewert S, Jones T, Mitchell M, Pitha-Rowe P, Freemont P, Taylor-Papadimitriou J. A novel gene (PLU-1) containing highly conserved putative DNA chromatin binding motifs is specifically up-regulated in breast cancer. Journal of Biological Chemistry. 1999;274:15633–15645. doi: 10.1074/jbc.274.22.15633. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–U791. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Mistell T. Hyperdynamic plasticity in pluripotent embryonic of chromatin proteins stem cells. Developmental Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–U552. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Tidow C, Klein HU, Hascher A, Isken F, Tickenbrock L, Thoennissen N, Agrawal-Singh S, Tschanter P, Disselhoff C, Wang YP, et al. Profiling of histone H3 lysine 9 trimethylation levels predicts transcription factor activity and survival in acute myeloid leukemia. Blood. 2010;116:3564–3571. doi: 10.1182/blood-2009-09-240978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin YH, Jiang Q, Li YQ, Coffield VM, Su LS, Xu GL, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Pedersen MT, Helin K. Histone demethylases in development and disease. Trends in Cell Biology. 2010;20:662–671. doi: 10.1016/j.tcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A Temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Mueller AM, Sterapfl T, Moehle C, Landthaler M, Vogt T. RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. International Journal of Cancer. 2008;122:1047–1057. doi: 10.1002/ijc.23211. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: Using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, Wang Q, Chia D, Goodglick L, Kurdistani SK. Global levels of histone modifications predict prognosis in different cancers. American Journal of Pathology. 2009;174:1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- Somervaille TCP, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Somervaille TCP, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, Chang HY, Shurtleff SA, Downing JR, Cleary ML. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun DQ, Luo M, Jeong M, Rodriguez B, Xia Z, Hannah R, Wang H, Le T, Faull KF, Chen R, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14:673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel AT, Blessington P, Zou T, Feather D, Wu XJ, Yan JZ, Zhang H, Liu ZG, Ernst P, Koretzky GA, Hua XX. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nature Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2 Elegant graphics for data analysis introduction. Ggplot2: Elegant Graphics for Data Analysis. 2009:1. + [Google Scholar]
- Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP, Chen CD. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. Embo Journal. 2011;30:1473–1484. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 dernethylase involved in transcriptional repression and breast cancer cell proliferation. Molecular Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Yamazaki J, Estecio MR, Lu Y, Long H, Malouf GG, Graber D, Huo YJ, Ramagli L, Liang SD, Kornblau SM, et al. The epigenome of AML stem and progenitor cells. Epigenetics. 2013;8:92–104. doi: 10.4161/epi.23243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye T, Krebs AR, Choukrallah M-A, Keime C, Plewniak F, Davidson I, Tora L. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Research. 2011;39 doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.