Summary
Lipid droplets (LD) are lipid storage organelles that grow or shrink, depending on the availability of metabolic energy. Proteins recruited to LDs mediate many metabolic functions, including phosphatidylcholine and triglyceride synthesis. How the LD protein composition is tuned to the supply and demand for lipids remains unclear. We show that LDs, in contrast to other organelles, have limited capacity for protein binding. Consequently, macromolecular crowding plays a major role in determining LD protein composition. During lipolysis, when LDs and their surfaces shrink, some, but not all, proteins become displaced. In vitro studies show that macromolecular crowding, rather than changes in monolayer lipid composition, causes proteins to fall off the LD surface. As predicted by a crowding model, proteins compete for binding to the surfaces of LDs. Moreover, the LD binding affinity determines protein localization during lipolysis. Our findings identify protein crowding as an important principle in determining LD protein composition.
Graphical abstract
Introduction
Most cells store neutral lipids, such as triglycerides (TGs) and sterol esters, in cytoplasmic organelles called lipid droplets (LDs) (Beller et al., 2010; Greenberg and Coleman, 2011; Walther and Farese, 2012). LDs are dynamic: their sizes depend on the metabolic state and therefore continually change. When lipids, such as fatty acids or sterols, are in excess, they are converted to neutral lipids and are stored in new or expanding LDs. Conversely, when cells require lipids for metabolic energy or membrane components, they catabolize neutral lipids from these organelles by lipolysis (Zanghellini et al., 2008; Zechner et al., 2009), resulting in LD shrinkage (Paar et al., 2012).
LDs are bounded by a surface monolayer, composed primarily of phospholipids and proteins. Many of these proteins mediate lipid metabolism (Athenstaedt et al., 1999; Brasaemle et al., 2004; Fujimoto et al., 2004; Krahmer et al., 2013; Pol et al., 2014). These include enzymes of TG synthesis (e.g., glycerol-3-phosphate acyltransferase 4 (GPAT4) and acyl CoA:diacylglycerol acyltransferase 2 (DGAT2) (Athenstaedt and Daum, 1997; Kuerschner et al., 2008; Stone et al., 2006; Wilfling et al., 2013), TG lipolysis (Grönke et al., 2005; Kurat et al., 2006; Zimmermann et al., 2004) (e.g., ATGL/brummer), and phosphatidylcholine (PC) synthesis (e.g., CTP-phosphocholine cytidylyltransferase; CCT). One of these enzymes, CCT, the rate-limiting enzyme for PC synthesis, is activated upon binding expanding LDs, catalyzing increased PC production for coating the growing LD surfaces (Krahmer et al., 2012). Other proteins targeted to the surfaces of LDs include important regulatory proteins, such as perilipin-adipophilin-TIP47 (PAT) proteins and LSD proteins in Drosophila (Brasaemle et al., 1997; Greenberg et al., 1991; Wolins et al., 2006; Wolins et al., 2001) and proteins that promote LD fusion (e.g., CIDE proteins (Gong et al., 2011; Jambunathan et al., 2011).
Proteins are targeted to the surface of LDs by at least two distinct mechanisms. Some proteins, including CCT, bind LDs by inserting their amphipathic helices into the surrounding phospholipid monolayer. These protein segments are likely disordered in the aqueous cytosol and become ordered upon binding to the LD surface (Bigay et al., 2005; Cui et al., 2011; Drin et al., 2007; Dunne et al., 1996; Thiam et al., 2013b). Other proteins, including GPAT4, DGAT2, and the putative lipase CG9186, (Goo et al., 2014; Thiel et al., 2013) localize to LDs using ER-LD bridges (Jacquier et al., 2011; Wilfling et al., 2013). The localization of each of these proteins is mediated by a hydrophobic, membrane-embedded domain that facilitates their delivery from the ER bilayer to the LD surface (Ingelmo-Torres et al., 2009; Wilfling et al., 2013; Zehmer et al., 2008).
Despite increased understanding of how proteins are targeted to the surface of LDs, the mechanisms that determine protein composition of LDs remain unclear. The targeting of some proteins, including hormone-sensitive lipase (HSL), ATGL, CGI-58, and CCT, is regulated by phosphorylation, which is dependent on the cellular metabolic state (Egan et al., 1992; Arnold et al., 1997; Brasaemle et al., 2000; Sahu-Osen et al., 2014; Xie et al., 2014). However, the principles regulating the relative amounts of these and other proteins at LD surfaces are not understood.
LDs possess unusual properties that necessitate distinct protein targeting mechanisms. For example, unlike other organelles bounded by bilayer membranes, LDs consist of a phospholipid monolayer surrounding a neutral lipid core. Therefore, the surface of LDs is unable to accommodate transmembrane proteins with hydrophilic luminal domains. Furthermore, in contrast to other large membranous organelles, such as the ER or Golgi, LDs are discrete entities with only limited binding surfaces. When LDs expand, their surface area increases, providing a platform for additional proteins to bind and mediate aspects of LD growth. For example, CCT normally resides in the nucleus or cytosol but specifically targets expanding LDs when excess fatty acids drive TG synthesis and storage (Krahmer et al., 2012). When LDs shrink during lipolysis, their binding surface decreases. How proteins are removed from LDs when they shrink is unknown.
Here, we investigated mechanisms that determine the protein composition of LDs. Using a combination of cell-based and in vitro reconstitution studies, we uncover macromolecular crowding as a major principle that mediates changes of protein composition of LDs. Our findings suggest that different binding affinities of proteins have evolved to fine-tune the LD protein composition to meet cellular needs.
Results
Lipid Droplet Protein Composition Changes During Lipolysis
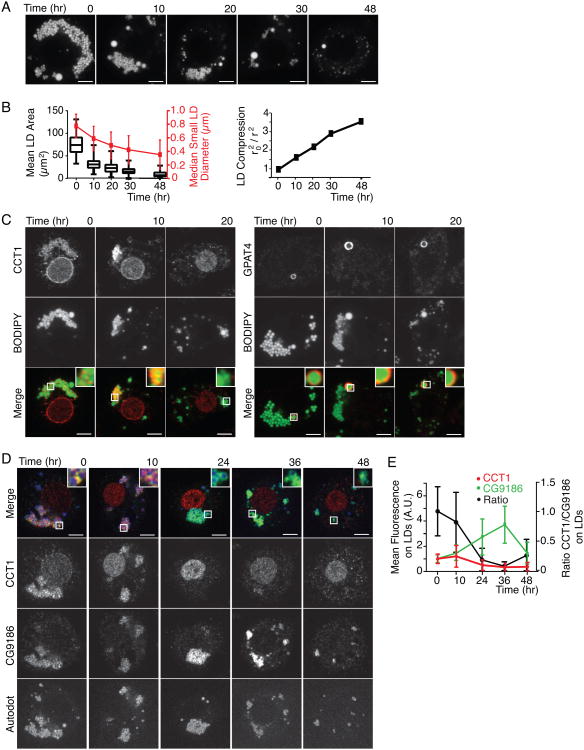
We first investigated the localization of LD proteins during lipolysis, which results in marked shrinkage of LD surfaces. To study this process, we incubated oleate-loaded Drosophila S2 cells in media lacking lipids, which leads to mobilization of their lipid stores. At the start of the experiment, cells had many LDs smaller than 1 μm in diameter (Figures 1A and 1B). After 48 hours of lipid deprivation, LDs were consumed or decreased dramatically in size (∼50% reduction in median diameter), resulting in a ∼3.5-fold compression of their surface areas (Figure 1B).
Figure 1. During Lipolysis, LDs Shrink and LD Protein Composition Changes.
(A) LDs are consumed during lipid starvation. After 48 hr in medium withoutlipids, LDs shrink and are removed from cells. LDs are stained with BODIPY.Representative images are shown. Scale bar, 5 μm.
(B) LD size decreases during lipid starvation. Mean LD area per cell, median LDdiameter of the small LD population, and compression factor (r2(time 0)/r2(respective time point); r = radius) during lipolysis is shown. Values are means± SD or medians as indicated (n > 20).
(C) Endogenous CCT1 detected by immunofluorescence, but not GPAT4, isdisplaced from LDs during shrinkage. Representative images are shown. Scalebar, 5 μm. Inlay, 3× magnification.
(D,E) Endogenous CCT1 is displaced from LDs during lipid starvation, whereas CG9186 concentrates on LDs. (D) LDs were stained with AUTOdot. Representative images are shown. Scale bar, 5 μm. Inlay, 3× magnification. (E) Mean fluorescence on LDs ± SD (n > 20). A.U. = arbitrary units. Note, CG9186 remained targeted to their surfaces throughout lipolysis, suggesting that these structures are cytosolic LDs.
We examined the localization of proteins during LD shrinkage by immunofluorescence, focusing on two proteins that are targeted to LDs by two distinct mechanisms: (1) CCT1, which binds LDs via an amphipathic helix, and (2) GPAT4, which binds via a hydrophobic hairpin motif (Krahmer et al., 2012; Wilfling et al., 2013). Endogenous CCT1 was present on LDs before and after 10 hr of lipid deprivation, but was almost completely absent from LDs after 20 hr, when instead it localized to the cell nucleus (Figure 1C, left panel). In contrast, endogenous GPAT4 remained on LDs (Figure 1C, right panel).
To determine whether the localization of other proteins changes during lipolysis, we co-stained the cells with antibodies against CCT1 and CG9186, a putative lipase (Goo et al., 2014; Thiel et al., 2013) during lipid starvation. Like GPAT4, CG9186 has a hydrophobic LD binding motif that is predicted to have hairpin structure (Thiel et al., 2013). Both CCT1 and CG9186 localized to the same LDs at the beginning of the time course (Figure 1D). As expected, CCT1 was no longer found on LDs between 10 and 24 hr of lipid deprivation (Figure 1D and (Krahmer et al., 2012)). In contrast, CG9186 increased in concentration three-fold after 30–36 hr of lipolysis and remained on LDs (Figures 1D and 1E). This increase correlated with a three-fold decrease in LD surface area (Figures 1B, 1E).
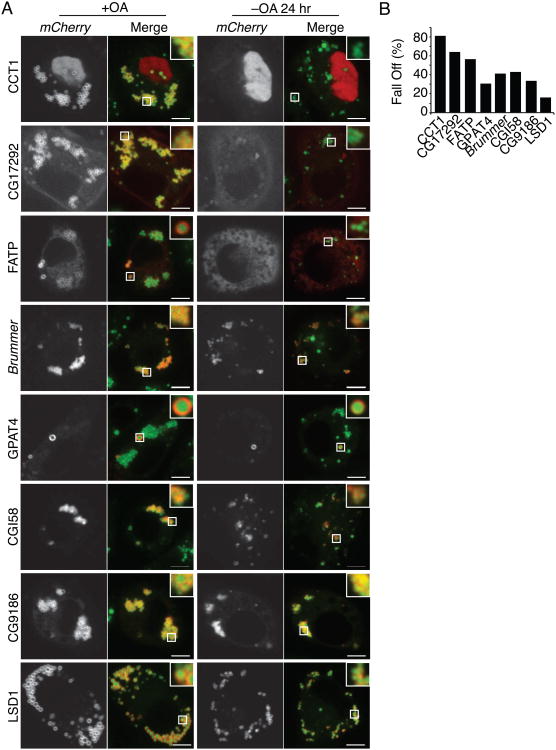
Next, we extended our analyses to a series of proteins that bind LDs by various mechanisms. These included proteins involved in lipolysis, including CG17292, ATGL, CGI-58, or TG synthesis, such as fatty acid transport protein (FATP). We expressed mCherry-tagged forms of these proteins and examined their localization during lipolysis. Each of the proteins localized to LDs at the beginning of the time course (Figure 2A). The binding of some of these proteins, such as CCT1 and CG17292, was strongly reduced after lipid deprivation (81% and 64% reductions, respectively, after 24 hr; Figures 2A and 2B). In contrast, other proteins, such as CG9186 and LSD1, remained mostly bound (34% and 16% reduction, respectively, after 24 hr; Figures 2A and 2B). In general, levels of amphipathic helix-containing proteins such as CCT1 and CG17292 were reduced on LDs during lipolysis, whereas levels of proteins with more hydrophobic LD-binding domains, such as GPAT4, CG9186, or multiple LD-binding motifs, such as LSD1 (Arrese et al., 2008), remained mostly bound (Figure 2B).
Figure 2. Differential Binding of LD Proteins during Lipolysis.
During lipolysis, some proteins are reduced on LDs whereas others remain bound. Cells were imaged after oleate loading (+OA) or after 24 hr (-OA 24 hr) of lipid starvation. LDs were stained with BODIPY. (A) Representative images are shown. Scale bar, 5 μm. Inlay, 3× magnification. (B) Percent protein displacement (% protein initially on LDs – % protein on LDs after starvation)/(% protein initially on LDs) is reported. Values are means (n > 12).
CCT1 Falls Off Shrinking Lipid Droplets, but Is Not Degraded
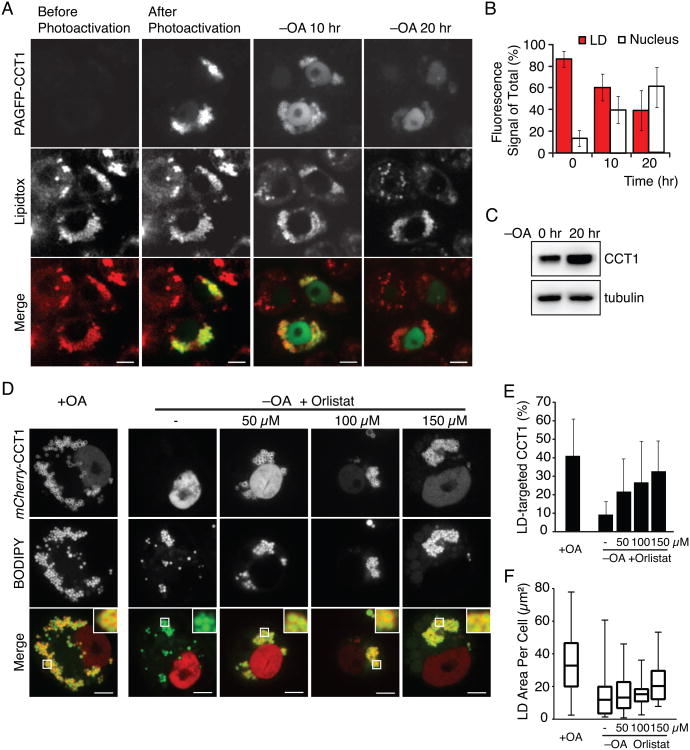
We reasoned that CCT is displaced from LDs during lipolysis. However, it is also possible that CCT1 is degraded during lipolysis and newly synthesized CCT1 subsequently is restricted to the nucleus. To rule out this possibility, we generated CCT1 fused to photo-activatable GFP and locally activated this protein at LDs during lipolysis (Patterson and Lippincott-Schwartz, 2002). During lipid deprivation and LD shrinkage, the pool of fluorescent CCT1 gradually disappeared from the LD surface and appeared in the nucleus (Figures 3A and 3B). In addition, the amount of photo-activated GFP-CCT1 protein was not reduced in the first 10 hr of starvation (data not shown), and total levels of the enzyme increased during 24 hr of starvation (Figure 3C). The results therefore suggest that CCT is displaced from LDs but is not degraded during lipolysis.
Figure 3. CCT1 Falls Off Shrinking LDs.
(A, B) CCT1 is not degraded but falls off LDs when cells are starved for lipids. Photoactivatable GFP (PAGFP)-CCT1 was activated on LDs before starvation. Cells were imaged before, immediately after photoactivation, and after 10 hr and 20 hr in medium containing delipidated serum (–OA 10 hr, –OA 20 hr, respectively). LDs were stained with LipidTOX. (A) Representative images are shown. Scale bar, 10 μm. (B) Percent mean fluorescence of PAGFP-CCT1 on LDs and the nucleus ± SD (n = 10).
(C) Total CCT1 levels increase during the first 20 hr of starvation. A representative Western blot using an antibody against endogenous CCT1 in cell lysates is shown. Tubulin was used as a loading control.
(D,E,F) Lipase inhibition blocks CCT1 displacement. Cells expressing mCherry-CCT1 were oleate loaded, imaged (+OA), or oleate loaded, starved of lipids for 24 hr in the presence of 0–150 μM Orlistat in DMSO and imaged (–OA). LDs were stained with BODIPY. (D) Representative images are shown. Scale bar = 5 μm. Inlay, 3× magnification. (E) Percent mean fluorescence of mCherry-CCT1 on LDs ± SD. Values are means (n > 12).
(F) Lipase inhibition prevents LD shrinkage and clearance. A box plot is shown. Mean values of the LD area in one plane of the cell are reported. Whiskers indicate Min and Max values. See also Figure S1.
CCT1 Displacement from Lipid Droplets Requires Lipolysis
We considered several mechanisms underlying the displacement of CCT1 from LDs during lipolysis. First, changes in the metabolic state during lipid starvation might activate enzymes that modify CCT1 (e.g. by phosphorylation), changing its binding affinity and localization. Second, changes in lipid composition at the LD surface due to the accumulation of lipid metabolites generated by lipolysis could re-localize CCT to the nucleus. Third, CCT1 could be crowded away from the shrinking LD surface.
To distinguish between these possibilities, we first tested whether lipolysis is required for the re-localization of CCT1 to the nucleus. Blocking lipolysis by the treatment with the lipase inhibitor Orlistat reduced LD shrinkage and prevented CCT1 re-localization (Figures 3D-F). This suggests that a change of properties at the LD surface, rather than posttranslational modification of CCT1 through the cell signaling of fatty acid starvation, is responsible for CCT1 release. If this is the case, we reasoned that a minimal LD-binding amphipathic helix motif of CCT1 (M-domain; Figure S1A; (Krahmer et al., 2012)), which is not known to be posttranslationally modified, would be sufficient to exhibit displacement from shrinking LDs. Indeed, we found that the M-domain of CCT1 was released from LDs at a similar rate as wild-type CCT1 during LD shrinkage (Figures S1B and S1C).
Surface Shrinkage Is Sufficient for Displacement of Some Lipid Droplet Proteins
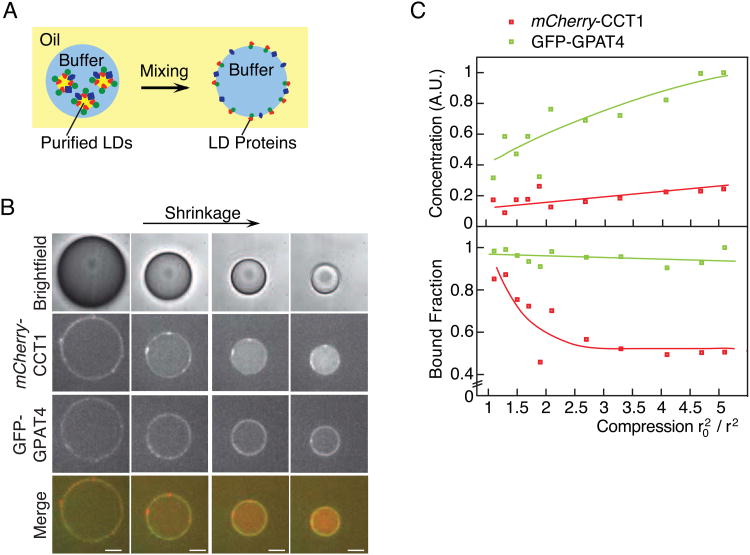
Our results suggest that shrinkage of LDs during lipolysis might be sufficient to preferentially displace some proteins from their surfaces. To evaluate this possibility, we developed an in vitro system using an oil-water interface that recapitulated monolayer shrinkage. We purified LDs from Drosophila S2 cells expressing fluorescently tagged LD proteins and mixed them in buffer with an excess of TG. In this system, LD proteins bind to the oil-water interface and can be analyzed by fluorescence microscopy (Figure 4A). Although this system creates an inverse emulsion, the opposite monolayer curvature is irrelevant because the size of these water-in-oil drops (>10-μm diameter), like the size of LD surfaces, is so large that the surface is considered flat on the molecular scale.
Figure 4. CCT1, but Not GPAT4, Falls Off a Shrinking Oil-Water Interface In Vitro.
(A) Schematic of the in vitro system. LDs in buffer are mixed with TG oil to generate a water-in-oil emulsion. LD proteins then bind to the resulting oil-water interface.
(B,C) During shrinkage of drops in vitro, CCT1 falls off the oil-water interface, whereas GPAT4 remains bound. (B) Representative images are shown. Scale bar, 10 μm. (C) Surface mean concentration and mean surface-bound fraction for mCherry-CCT1 and GFP-GPAT4 are reported. Lines represent trends. A.U. = arbitrary units. See also Figure S2.
Importantly, because the oil phase is experimentally accessible, the influence of different factors at the interface, such as phospholipid concentration, can be tested. To simulate shrinkage of the surface of LDs during lipolysis, water can be evaporated over time from the aqueous drops by adjusting the humidity, leading to shrinkage of the oil-water surface. During shrinkage, the volume of the oil phase remains constant and equilibration of phospholipids between the oil phase and the surface maintains the monolayer lipid composition, allowing the effects of macromolecular crowding from effects of changing surface lipid composition to be independently evaluated.
Using this in vitro system, we evaluated whether shrinkage alone could displace CCT1 from the oil-water interface. We found that during drop shrinkage the CCT1 signal decreased from the interface and concomitantly increased in the aqueous phase (Figures 4B and 4C). In contrast, proteins that stay on shrinking cellular LDs, such as GPAT4, LSD1, or CG9186 (Figures 4 and S2) remained at the oil-water interface and increased in concentration as the surface shrunk. No changes, other than surface shrinkage, were required to recapitulate the displacement of LD proteins from the interface in the in vitro system.
Changes in the Composition of Surface Lipids at Oil-water Interfaces Are not Sufficient to Displace Proteins
It is possible that CCT1 might fall off LDs during lipolysis due to changes in lipid composition at the shrinking LD surface. Indeed, during LD expansion, when levels of PC are reduced, CCT1 binds LDs (Krahmer et al., 2012). We therefore tested if increasing the concentration of PC at the shrinking oil-water interface is sufficient to displace CCT1 from the oil-water interface. To test this possibility, we added 25 mM PC to the oil phase, a concentration vastly exceeding its critical micellar concentration in oil (∼ 0.5 mM) or water (nanomolar). This leads to saturation of the oil-water interface, with excess PC predominantly partitioninginto the oil phase. Under this condition, and in the absence of drop shrinkage, CCT1 remained bound to the oil-water interface (Figures 5A and 5B). Similarly, adding other lipids to the interface, including either fatty acids, diacylglycerol, monoacylglycerol, a phosphatidylethanolamine(PE)-PC mixture, or a phospholipid mixture mimicking the LD surface composition, did not reduce the amount of CCT1 bound to oil-water interface (Figure S3A). Furthermore, we confirmed that added phospholipids reached the oil-buffer interface (data not shown) by addition of the fluorescent tracer, rhodamine-PE (data not shown). These results indicate that changes in the lipid composition of the interface lipids alone are insufficient to affect binding of CCT1 to the oil-water interface in the in vitro system.
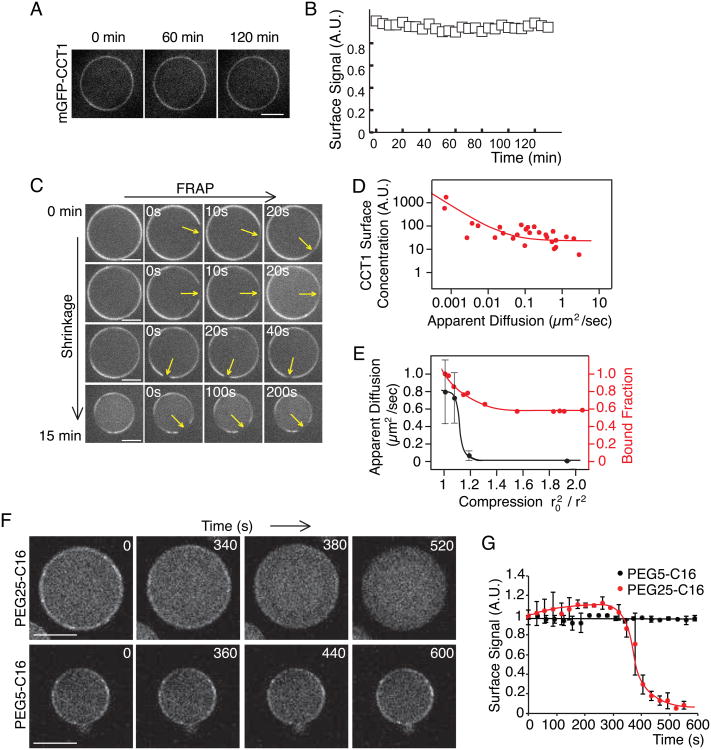
Figure 5. Macromolecular Crowding, Not Changes in PC Concentration, Causes CCT1 Displacement In Vitro.
(A,B) PC addition does not affect CCT1 binding to the oil-water interface. Excess PC (2% w/w to TG, 25mM) was added to the TG oil phase of the inverse emulsion after mGFP-CCT1 was bound at the oil-water interface. (A) Representative images are shown. Scale bar, 10 μm. (B) Mean fluorescence on LDs ± SD (n = 11). A.U. = arbitrary units.
(C) Protein diffusion at the oil-water interface of an in vitro drop is gradually decreased upon interface shrinkage according to FRAP analysis. Representative images are shown. Scale bar, 10 μm.
(D) The diffusion of CCT at the oil-water interface is inversely correlated with the concentration of CCT at the drop surface according to FRAP analysis (C). D ∞ 1/C, assuming a Stoke-Einstein-like law, is used to fit the data. Note, shrunken drops have a high concentration of CCT1 at their surface and volume and a low diffusion rate along the surface. A.U. = arbitrary units.
(E) CCT1 displacement occurs before its diffusion is limited. Mean diffusion (±SD, n = 4) and fraction of surface-bound CCT1 were measured on drops andplotted against the compression factor of the drop. Lines indicate trends.
(F,G) High-, but not low-, molecular-weight PEGs crowd out CCT1 from the oil-water interface. PEGs were added at room temperature (2% w/w of the oil) to drops whose interface was bound by mGFP-CCT1. (F) Representative images are shown. Scale bar, 50 μm. (G) Mean mGFP-CCT1 fluorescence ± SD (n = 7) on the drop surface for indicated times over time is shown. The value at time 0 was normalized to 1. Lines are trend lines. A.U. = arbitrary units. See also Figure S3.
Macromolecular Crowding Mediates Protein Displacement from Shrinking Oil-Water Interfaces
Our results suggest that during shrinkage, LD proteins become crowded at the surface, displacing weakly associated proteins. To test whether the oil-water interface indeed becomes crowded during shrinkage, we used fluorescence-recovery-after-photobleaching (FRAP) to measure the lateral diffusion of proteins on the oil-water interphase before and during drop shrinkage. A slowing of diffusion is the hallmark of macromolecular crowding (Frick et al., 2007; Goose and Sansom, 2013; Han and Herzfeld, 1993; Zimmerman and Minton, 1993). We found that mCherry-CCT1 diffused laterally along the interface (Figure 5C). However, under conditions of interface shrinkage, the diffusion rate was dramatically reduced. Importantly, the diffusion rate was inversely correlated with the surface compression factor, with almost no diffusion occurring at a compression factor ≥ 2 (Figures 5C-E). At extreme compression, the high density of protein led to buckling of the interface (Figure S3B).
We reasoned that if macromolecular crowding is responsible for the release of some proteins, such as CCT1, from shrinking oil-water interfaces, the addition of high-molecular-weight polyethylene glycol (PEG), a crowding agent, should produce similar effects. To assess this possibility, we added PEG conjugated to C-16 fatty alcohols in our in vitro system. PEG–fatty acid conjugates are widely used for binding oil-water interfaces to stabilize emulsions, and are known to diffuse to these interfaces to fully cover them (Wheeler et al., 1994). As predicted, adding PEG25-C16 displaced CCT1 from the oil-water-interface without drop shrinkage at room temperature (Figures 5F and 5G). In contrast, adding a smaller molecule, PEG5-C16, at the same concentration had no effect.
Proteins Compete for Binding the Lipid Droplet Surface
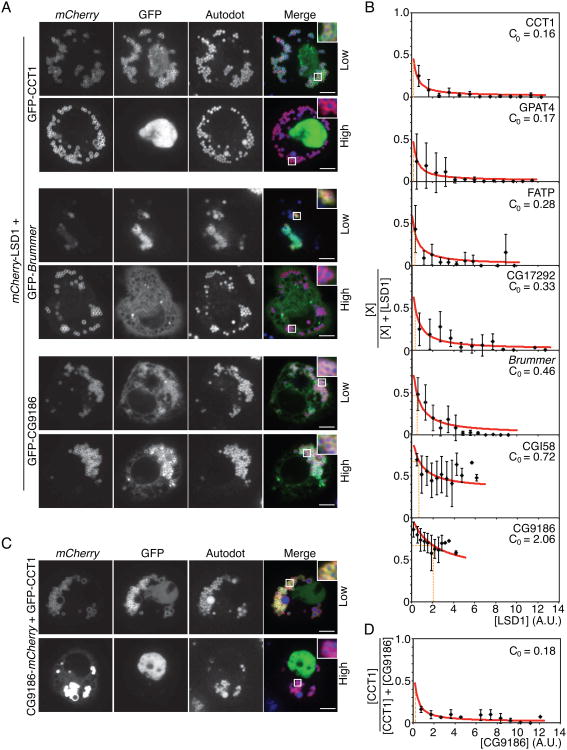
If crowding displaces weakly associated proteins from the shrinking surface during lipolysis, we hypothesized that increasing levels of a protein with high LD binding affinity would change the LD protein composition at a steady state. To test this prediction, we established competition assays in Drosophila S2 cells under conditions in which LDs are abundant at a relatively steady state. In brief, we co-expressed a series of LD proteins, together with LSD1, in Drosophila S2 cells. Under these conditions, mCherry-LSD1 was predominantly localized to LDs in all experiments (80% when co-expressed with CCT1; Figures 6A and S4D). In contrast, increased levels of mCherry-LSD1 resulted in decreased levels of most GFP-tagged proteins on the surface of LDs (Figure 6A, Figure S4A). However, some proteins, such as CG9186 (Figure 6A) and CGI-58 (Figures 6A and S4A), were unaffected.
Figure 6. Proteins Compete for Binding at the Lipid Droplet Surface.
(A) High levels of LSD1 compete off some, but not all, LD proteins. mCherry-LSD1 was co-expressed with GFP-CCT1 in LD-containing Drosophila S2 cells. One representative cell with low expression (upper panel) and one with high expression of LSD1 (lower panel) are shown. LDs were stained with AUTOdot. Scale bar is 5 μm. Inlay, 3× magnification.
(B) Some proteins compete more strongly than others against LSD1 at the LD binding surface. Mean fluorescence on LDs ± SD (n > 15). A.U. = arbitrary units.
(C,D) High levels of CG9186 outcompete CCT at the surface of LDs. mCherry-CG9186 was co-expressed with GFP-CCT1 in LD-containing Drosophila S2 cells. (C) One representative cell with low expression (upper panel) and one with high expression of LSD1 (lower panel) are shown. LDs were stained with AUTOdot. Scale bar is 5 μm. Inlay, (D) Mean fluorescence on LDs ± SD (n > 15). A.U. = arbitrary units. See also Figure S4.
Using these results, we estimated the relative binding affinities of different proteins for the surface of LDs (see Experimental Procedures). Among the proteins tested, CCT1 was most easily displaced by LSD1 (C0=0.16), followed by GPAT4 (C0=0.17), FATP (C0=0.28), CG17292 (C0=0.33), and brummer (C0=0.46). CGI58 (C0=0.72) and CG9186 (C0=2.06) had the strongest affinity for the LD surface, compared with LSD1, and were not displaced from LDs even at the highest concentration of LSD1 (Figures 6B and S4A).
To further confirm these results, we performed a similar analysis using a mCherry-tagged form of the putative lipase CG9186 as a reference. These experiments yielded similar results (Figures 6C and 6D; Figures S4B and S4C). Importantly, at high expression levels, CG9186, like LSD1, displaced CCT1 and CG17292 from LDs. These results, in combination with the relative increase in concentration of CG9186 during lipolysis (Figures 1D and 1E), supports the hypothesis that increased crowding at the LD surface is responsible for displacement of CCT1 and other proteins during lipolysis.
Lipid Droplet Binding Affinity Determines Localization During Lipolysis
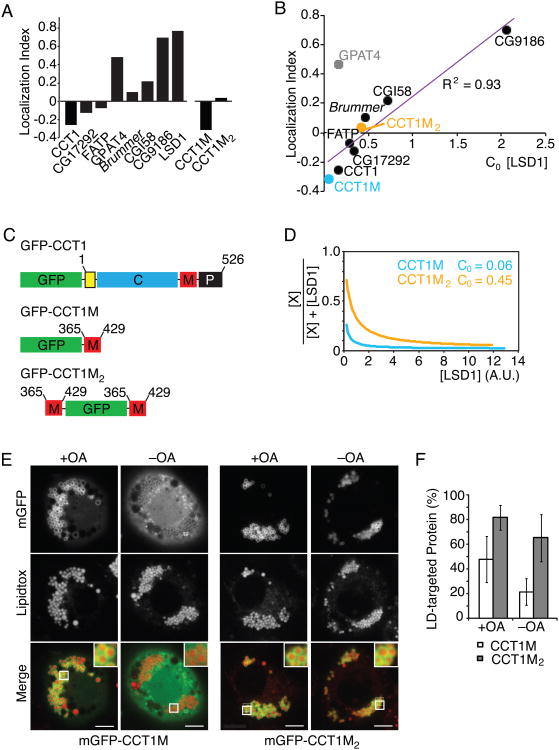
If competition for the shrinking LD surface is a key determinant for LD protein composition during lipolysis, we hypothesized that the degree of displacement would inversely correlate with binding affinities at steady state. To evaluate this possibility, we defined a localization index for each protein. To calculate this index, we first compared the percentage of a protein on LDs with that elsewhere in the cell (Figure 2B) and normalized this ratio to the LD area to correct for effects of protein overexpression on LD abundance. Next, we calculated the fold change of protein on LDs after shrinkage compared with before lipid deprivation. The localization index is defined as the difference of the fold change from 1 (Figure 7A). Among the proteins analyzed, CCT1, CG17292, and FATP were reduced in concentration on LDs during shrinkage, reflected in a negative localization index. In contrast, ATGL, GPAT4, CGI58, CG9186, and LSD1 increased in LD concentration, reflected in a positive localization index (Figure 7A).
Figure 7. Binding Affinity Determines Protein Lipid Droplet Localization During Lipolysis.
(A) LD proteins are displaced from LDs to different degrees during lipid starvation. The localization index is defined as the difference of the fold change in percentage of a protein on LDs versus the rest of the cell from 1.
(B) The correlation of localization index and critical LSD1 concentration needed to replace half of the amount of a bound protein from LDs as determined in Figure 6B. Linear regression, GPAT4 data was omitted from modeling.
(C) A schematic of GFP-tagged full-length CCT1, the LD binding domain (M-domain) and two copies of the M domain.
(D) LSD1 displaces mGFP-CCT1M at a lower concentration than mGFP-CCT1M2. A.U. = arbitrary units.
(E,F) mGFP-CCT1M2 falls off LDs less than mGFP-CCT1M. LDs were stained with LipidTOX. (E) Representative images are shown. Scale bar, 5 μm. Inlay, 3× magnification. (F) Mean fluorescence on LDs ± SD (n > 12). A.U. = arbitrary units. See also Figure S5.
For almost every protein tested, the localization index correlated strongly with the ability of each protein to compete for LD binding surface at steady state (Figure 7B). This suggests that the same fundamental principle – competition for limited binding sites on a crowded surface – underlies both protein displacement and LD localization at steady-state. One exception is GPAT4, which was easily displaced by LSD1 but mostly remained bound after 24 h of lipid deprivation. The explanation for this exception is currently unclear. We showed previously that GPAT4 is targeted to LDs via membrane bridges through an Arf1/COPI-dependent mechanism (Wilfling et al., 2014; Wilfling et al., 2013). It is therefore possible that LSD1 displaces targeting factors required for GPAT4 localization such as components of the Arf1/COPI machinery.
Our results suggest a model in which the binding of proteins to LDs is determined by their affinity for the LD surface, and that weakly associated proteins become displaced during lipolysis. To further test this idea, we increased the affinity of CCT LD-binding domain by fusing two copies of this domain (GFP-CCT1M2; Figure 7C) and tested its behavior during lipolysis. As predicted, this construct has a higher affinity for the LD surface than one with a single M-domain according to FRAP analysis (estimated on-rate: 0.047/min versus 1.13/min for the single M-domain, Figures S5A and S5B). Furthermore, when co-expressed with LSD1, GFP-CCT1M2 competed more efficiently for binding than the single M-domain (Figure 7D). To test whether this change in affinity leads to increased binding to the LD surface, we investigated the localization of both constructs during lipolysis. As predicted, GFP-CCT1M2 remained on LDs to a greater extent than the single M-domain fusion (65% of the signal vs. 20%) (Figures 7E and 7F). In addition, for both M-domain constructs, the localization index correlated with their binding affinity (Figure 7B).
Discussion
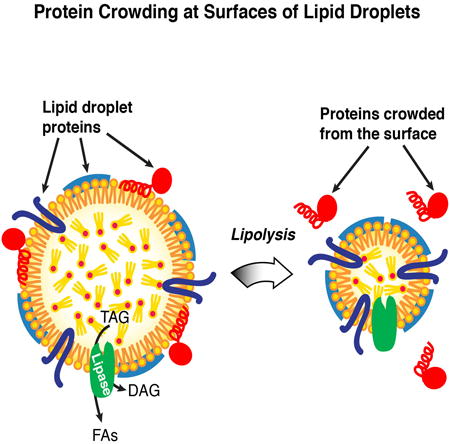
Here, we show that macromolecular crowding is a major determinant of LD protein composition. During lipolysis, protein crowding alters LD composition by gradually expelling proteins from their shrinking surfaces according to binding strength. Our in vitro studies show that this displacement occurs due to macromolecular crowding at the oil-water interface. Furthermore, when LD surfaces are at steady state, increasing levels of proteins with high LD binding affinity changes the LD protein composition, suggesting competition between proteins for the binding surface. Taken together, our results reveal a mechanism that governs the relative amounts of different LD proteins as they expand or contract.
The mechanisms regulating protein composition of LDs is apparently different from those that govern the composition of other membrane organelles, such as the ER, Golgi, or mitochondria. In the latter cases, protein composition is determined largely by expression and degradation of proteins, with signal sequences allowing import of proteins to the organelle (Nunnari and Walter, 1996). Alternatively, interactions of specific protein domains with highly enriched membrane lipid determinants, such as phosphoinositides, recruit proteins to these organelles. However, no such LD-specific determinants have been identified. Instead, we propose that the protein composition of LDs is determined in large part by competition for binding to limited sites on the monolayer surface. Because LDs exist as the dispersed oil phase of cellular emulsions, the available LD surface of individual droplets is coupled to the abundance of neutral lipids, limiting the possibilities for volume regulation that can occur with other organelles.
Macromolecular crowding is an important cellular phenomenon, influencing the behavior of bilayer membranes. At the plasma membrane, asymmetric protein crowding leads to membrane bending to release the lateral pressure (Derganc et al., 2013; Stachowiak et al., 2010; Stachowiak et al., 2012). The effects of crowding differ in the case of LDs, where a surfactant monolayer covering a hydrophobic phase is more difficult to deform than a bilayer membrane (Thiam et al., 2013b). In this situation, lateral pressure from crowding leads to displacement of proteins rather than bending of the surface. This response to crowding is similar to findings that were reported for surface proteins of plasma lipoproteins (Mitsche and Small, 2013).
From our study, two classes of LD proteins emerge with respect to the effects of crowding. One class, which includes CCT1, targets LDs from the cytoplasm and binds to LDs by inserting amphipathic helices into the surrounding monolayer. These proteins are the most susceptible to displacement due to crowding at the LD surface. A second class includes proteins with more hydrophobic helices that insert into the ER and subsequently re-localize to forming or expanding LDs (Ingelmo-Torres et al., 2009; Jacquier et al., 2011; Wilfling et al., 2013; Zehmer et al., 2008). Generally, these hydrophobic proteins have higher LD binding affinities and are not crowded away from shrinking LDs during lipolysis. How cells remove these proteins from LDs when the droplets are entirely consumed is unclear.
Changes in monolayer lipid composition during lipolysis do not appear to contribute to displacement of weakly bound proteins, at least for CCT1. This contrasts with the binding of CCT1 to LDs, which is sensitive to PC deficiency and occurs during expansion to facilitate LD growth (Krahmer et al., 2012). Molecularly, the different sensitivity of CCT1 to surface lipids for binding versus displacement might be explained by the coupling of lipid binding to helix folding. When CCT binds to LDs, the folding of the amphipathic helical minimizes the energy penalty incurred by polar atoms being exposed to the hydrophobic environment of lipid side-chains. This step essentially renders the pathway of the binding reaction irreversible under these conditions (Antonny, 2011; Clayton et al., 2003). Therefore, CCT1 remains bound to the LD surface until proteins crowd, which increases collision events and causes its displacement from the surface.
Why some proteins are more easily displaced from the surface of LDs than others during crowding is an open question. One possibility is that the binding affinities of proteins targeted to LDs evolved due to selection pressures reflecting their functions in lipid storage or utilization. CCT1 provides an example. Previously, we showed that CCT1 exhibits a high apparent on-rate and binds tightly during LD expansion (Krahmer et al., 2012). Here, we show during lipolysis that CCT1 has a high propensity to fall off when LDs shrink, and CCT1 activity is no longer required. These properties reflect the need for CCT1 at the LD surface to provide PC during LD expansion, but not during LD shrinkage, when phospholipids are in excess, and CCT1 activity is no longer required. Other proteins, such as lipase co-activator CGI-58 or the putative lipase CG9186, have a much lower propensity to be displaced by crowding, ensuring they stay on LDs during lipolysis. Such a mechanism for lipases may facilitate metabolic energy generation by optimizing substrate access during continued LD shrinkage.
Our findings do not exclude that processes other than crowding regulate LD composition. For example, the binding of some proteins, such as HSL, ATGL, CGI58, and CCT1, is regulated by protein phosphorylation depending on metabolic state (Egan et al., 1992; Arnold et al., 1997; Brasaemle et al., 2000; Sahu-Osen et al., 2014; Xie et al., 2014). In addition, during LD expansion, surface lipid composition of LDs, such as deficiency of PC, influences the binding of CCT, and possibly of other LD proteins, to LDs to facilitate growth (Arnold et al., 1997; Jamil and Vance, 1990; Krahmer et al., 2012; Sletten et al., 2014). These mechanisms likely represent other layers of regulation that work in concert with protein crowding to control LD protein composition.
In conclusion, we propose that the unusual organelle structure of LDs – a monolayer interface and limited surface area – result in protein crowding serving as a general mechanism that determines their protein composition. As a mechanism, protein crowding may be advantageous to cells, as it enables the regulation of protein composition at the LD surface under changing conditions. For example, protein crowding may govern which proteins bind to LD surfaces during LD expansion versus shrinkage. According to this model, protein crowding would prevent proteins with weak affinities for membrane surfaces from binding to LDs during expansion. In this respect, PAT proteins, putative regulatory proteins found on most mammalian LDs, might serve such a crowding-related regulatory function. As we demonstrate, the PAT protein LSD1 has a high binding affinity for LDs and is efficient in competing other proteins off the LD surface. PAT proteins might therefore increase the stringency of proteins binding to LDs, effectively limiting binding to those proteins with relatively high affinity, thereby regulating the LD protein composition through a type of molecular proofreading.
Experimental Procedures
Cell Culture and Transfection
Drosophila S2 cell culture and LD inductions were performed as described previously (Wilfling et al., 2013). For lipid starvation experiments, cells were treated with oleic acid overnight, washed in PBS three times, and incubated in media supplemented with 5% delipidated FBS (Gemini Bio-Products, West Sacramento, CA). The medium was changed after 10, 24, and 32 hr. Lipolysis was blocked using the broad-specificity lipase inhibitor Orlistat (Cayman Chemical Company, Ann Arbor, MI).
Fluorescence Microscopy
Immunofluorescence and spinning-disk confocal microscopy (100 × 1.4 NA oil immersion objective [Olympus], iMIC [Till], CSU22 [Yokugawa], iXonEM 897 [Andor]) were performed as previously described (Wilfling et al., 2013). Primary antibodies against Drosophila CCT1, GPAT4 (Wilfling et al., 2013) or CG9186 (Haas et al., 2012) and fluorescently labeled secondary antibodies (Life Technologies, Grand Island, NY) were used. FRAP experiments were performed as described (Krahmer et al., 2012).
For coexpression competition experiments, mCherry- or GFP-tagged LD protein constructs in equal concentrations were transfected into S2 cells. After oleic acid treatment, cells expressing both proteins at various levels were imaged.
Image Quantification and Statistics
Images were analyzed using ImageJ software (Schneider et al., 2012). To determine the size of LDs, the diameters of the 20 largest LDs in one plane of the cell were measured. Small LDs were defined as less than 1.3 μm in size. To determine the LD area in one plane of the cell Otsu thresholding was applied to the BODIPY channel and the resulting area was measured. For quantification of the %LD-targeted signal for a given protein, the image was background-corrected and the total fluorescent signal on LDs was determined as a ratio to the total fluorescent signal in the whole cell. In co-expression experiments, the fluorescence signal on LDs was calculated by subtracting out the fluorescence signal elsewhere in each cell. Protein concentrations on LDs were derived from the mean fluorescence measured on LDs in each channel. Values from 15-20 cells were combined and the standard deviation was calculated for statistical analysis.
Photoactivation Experiment
PAGFP-CCT1 (Patterson and Lippincott-Schwartz, 2002) was activated on LDs in a number of cells and imaged before and after 10 hr or 20 hr of lipid starvation. The integrated signal on LDs and the nucleus from 10 cells were combined for statistical analysis.
Curve Fittings
For the co-expression experiments, we determined the concentration of the protein based on the mean fluorescence intensity, Prot, and the concentration of the reference protein, Protref (e.g. LSD1). To determine the fraction of displaced protein, we plotted Prot/(Prot+Protref) against Protref and fitted curves based on the function 1/(1+x/c0) to the binned data, where x is the variable Protref and C0 the concentration of mCherry-LSD1 at which half of the GFP-tagged protein is displaced from LDs (Protref = c0). This fitting model is based on the Stoke-Einstein equation: when the protein concentration is increased, the viscosity of the surface increased, which leads to impaired diffusion (diffusion D is inversely proportional to the viscosity). Since the amount of protein displacement correlated with surface diffusion (Figure 5E), we considered our fitting model adequate.
For the determination of the diffusion coefficient, we bleached part of the interface, in the in vivo experiments, of characteristic size l2, and determined the characteristic recovery time t. The diffusion coefficient was estimated as l2/t.
In Vitro Experiments
To purify LDs from cells expressing fluorescently-tagged LD proteins, cells from 3-5 10-cm dishes were harvested, washed once in ice-cold phosphate-buffered saline and lysed using a 30G needle. To isolate LDs, cell lysates were mixed with 1 ml of 75% glycerol in Tris-buffered saline (TBS) buffer, overlaid with 1.5 ml of TBS and spun at 100,000 × g for 1 h. The top 750 μl were collected as the LD fraction. To create buffer-in-oil drops, a buffer-diluted LD fraction was mixed with triacylglycerol by vortexing to create buffer-in-oil drops.
For shrinking experiments, aqueous drops bounded by the triacylglycerol were imaged for 10 to 15 min on uncovered glass plates to allow for water evaporation. Where indicated, lipids (Avanti Polar Lipids, Alabaster, AL) or PEG-C16 (TCI America, Boston, MA) conjugates were added to the oil phase on the coverslips at 0.5% and 2% w/w respectively where indicated. Surfactant lipids were first dried under vacuum before being resuspended.
Supplementary Material
Acknowledgments
We thank Drs. Matthias Beller for the CG9186 antibody, Natalie Krahmer and Florian Wilfling for constructs, and Yi Guo for mCherry-LSD1. We also thank Drs. Thomas Melia, Niklas Mejhert, and Coline Prévost for comments and critical discussion, and Dr. Michelle Pflumm and Gary Howard for editorial assistance. This work was supported by the Mathers foundation (T.C.W), the National Institute of General Medical Sciences (NIGMS) (R01GM-097194, T.C.W; R01GM-099844, R.V.F.) and the ATIP-Avenir program (A.R.T.). In addition, A.R.T held a Marie Curie Fellowship and N.K., an American Heart Association pre-doctoral fellowship.
Footnotes
Author Contributions: N.K., A.R.T., and T.C.W. designed the research; N.K. and A.R.T. performed research; N.K. and A.R.T. analyzed data; N.K., A.R.T., R.V.F. and T.C.W. interpreted results and N.K., R.V.F., and T.C.W. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonny B. Mechanisms of membrane curvature sensing. Annu Rev Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- Arnold RS, DePaoli-Roach AA, Cornell RB. Binding of CTP:phosphocholine cytidylyltransferase to lipid vesicles: diacylglycerol and enzyme dephosphorylation increase the affinity for negatively charged membranes. Biochemistry. 1997;36:6149–6156. doi: 10.1021/bi970023z. [DOI] [PubMed] [Google Scholar]
- Athenstaedt K, Daum G. Biosynthesis of phosphatidic acid in lipid particles and endoplasmic reticulum of Saccharomyces cerevisiae. J Bacteriol. 1997;179:7611–7616. doi: 10.1128/jb.179.24.7611-7616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K, Zweytick D, Jandrositz A, Kohlwein SD, Daum G. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J Bacteriol. 1999;181:6441–6448. doi: 10.1128/jb.181.20.6441-6448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller M, Thiel K, Thul PJ, Jäckle H. Lipid droplets: a dynamic organelle moves into focus. FEBS Lett. 2010;584:2176–2182. doi: 10.1016/j.febslet.2010.03.022. [DOI] [PubMed] [Google Scholar]
- Bigay J, Casella JF, Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasaemle DL, Barber T, Wolins NE, Serrero G, Blanchette-Mackie EJ, Londos C. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J Lipid Res. 1997;38:2249–2263. [PubMed] [Google Scholar]
- Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Levin DM, Adler-Wailes DC, Londos C. The lipolytic stimulation of 3T3-L1 adipocytes promotes the translocation of cytosolic hormone-sensitive lipase to the surfaces of lipid storage droplets. Biochim Biophys Acta. 2000;1483:251–262. doi: 10.1016/s1388-1981(99)00179-1. [DOI] [PubMed] [Google Scholar]
- Clayton AH, Vultureanu AG, Sawyer WH. Unfolding of class A amphipathic peptides on a lipid surface. Biochemistry. 2003;42:1747–1753. doi: 10.1021/bi020439z. [DOI] [PubMed] [Google Scholar]
- Cui H, Lyman E, Voth GA. Mechanism of membrane curvature sensing by amphipathic helix containing proteins. Biophys J. 2011;100:1271–1279. doi: 10.1016/j.bpj.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derganc J, Antonny B, Copic A. Membrane bending: the power of protein imbalance. Trends Biochem Sci. 2013;38:576–584. doi: 10.1016/j.tibs.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- Dunne SJ, Cornell RB, Johnson JE, Glover NR, Tracey AS. Structure of the membrane binding domain of CTP:phosphocholine cytidylyltransferase. Biochemistry. 1996;35:11975–11984. doi: 10.1021/bi960821+. [DOI] [PubMed] [Google Scholar]
- Egan JJ, Greenberg AS, Chang MK, Wek SA, Moos MC, Jr, Londos C. Mechanism of hormone-stimulated lipolysis in adipocytes: translocation of hormone-sensitive lipase to the lipid storage droplet. Proc Natl Acad Sci U S A. 1992;89:8537–8541. doi: 10.1073/pnas.89.18.8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick M, Schmidt K, Nichols BJ. Modulation of lateral diffusion in the plasma membrane by protein density. Curr Biol. 2007;17:462–467. doi: 10.1016/j.cub.2007.01.069. [DOI] [PubMed] [Google Scholar]
- Fujimoto Y, Itabe H, Sakai J, Makita M, Noda J, Mori M, Higashi Y, Kojima S, Takano T. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta. 2004;1644:47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Gong J, Sun Z, Wu L, Xu W, Schieber N, Xu D, Shui G, Yang H, Parton RG, Li P. Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J Cell Biol. 2011;195:953–963. doi: 10.1083/jcb.201104142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo YH, Son SH, Kreienberg PB, Paul A. Novel lipid droplet-associated serine hydrolase regulates macrophage cholesterol mobilization. Arterioscler Thromb Vasc Biol. 2014;34:386–396. doi: 10.1161/ATVBAHA.113.302448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goose JE, Sansom MS. Reduced lateral mobility of lipids and proteins in crowded membranes. PLoS Comput Biol. 2013;9:e1003033. doi: 10.1371/journal.pcbi.1003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AS, Coleman RA. Expanding roles for lipid droplets. Trends Endocrinol Metab. 2011;22:195–196. doi: 10.1016/j.tem.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchette-Mackie EJ, Londos C. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem. 1991;266:11341–11346. [PubMed] [Google Scholar]
- Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, Müller G, Jäckle H, Kühnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Haas JT, Winter HS, Lim E, Kirby A, Blumenstiel B, DeFelice M, Gabriel S, Jalas C, Branski D, Grueter CA, et al. DGAT1 mutation is linked to a congenital diarrheal disorder. The Journal of clinical investigation. 2012;122:4680–4684. doi: 10.1172/JCI64873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Herzfeld J. Macromolecular diffusion in crowded solutions. Biophys J. 1993;65:1155–1161. doi: 10.1016/S0006-3495(93)81145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelmo-Torres M, Gonzalez-Moreno E, Kassan A, Hanzal-Bayer M, Tebar F, Herms A, Grewal T, Hancock JF, Enrich C, Bosch M, et al. Hydrophobic and basic domains target proteins to lipid droplets. Traffic. 2009;10:1785–1801. doi: 10.1111/j.1600-0854.2009.00994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci. 2011;124:2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- Jambunathan S, Yin J, Khan W, Tamori Y, Puri V. FSP27 promotes lipid droplet clustering and then fusion to regulate triglyceride accumulation. PLoS One. 2011;6:e28614. doi: 10.1371/journal.pone.0028614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamil H, Vance DE. Head-group specificity for feedback regulation of CTP:phosphocholine cytidylyltransferase. Biochem J. 1990;270:749–754. doi: 10.1042/bj2700749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt-Supprian M, Vance DE, Mann M, et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2012;14:504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Hilger M, Kory N, Wilfling F, Stoehr G, Mann M, Farese RV, Jr, Walther TC. Protein correlation profiles identify lipid droplet proteins with high confidence. Molecular & cellular proteomics : MCP. 2013;12:1115–1126. doi: 10.1074/mcp.M112.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuerschner L, Moessinger C, Thiele C. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 2008;9:338–352. doi: 10.1111/j.1600-0854.2007.00689.x. [DOI] [PubMed] [Google Scholar]
- Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, Scholz H, Zimmermann R, Leber R, Zechner R, Kohlwein SD. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem. 2006;281:491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- Lass A, Zimmermann R, Oberer M, Zechner R. Lipolysis - a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog Lipid Res. 2011;50:14–27. doi: 10.1016/j.plipres.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsche MA, Small DM. Surface pressure-dependent conformation change of apolipoprotein-derived amphipathic alpha-helices. J Lipid Res. 2013;54:1578–1588. doi: 10.1194/jlr.M034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Walter P. Regulation of organelle biogenesis. Cell. 1996;84:89–94. doi: 10.1016/s0092-8674(00)81283-0. [DOI] [PubMed] [Google Scholar]
- Paar M, Jüngst C, Steiner NA, Magnes C, Sinner F, Kolb D, Lass A, Zimmermann R, Zumbusch A, Kohlwein SD, et al. Remodeling of lipid droplets during lipolysis and growth in adipocytes. J Biol Chem. 2012;287:11164–11173. doi: 10.1074/jbc.M111.316794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Pol A, Gross SP, Parton RG. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol. 2014;204:635–646. doi: 10.1083/jcb.201311051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu-Osen A, Montero-Moran G, Schittmayer M, Fritz K, Dinh A, Chang YF, McMahon D, Boeszoermenyi A, Cornaciu I, Russell D, et al. CGI-58/ABHD5 is phosphorylated on Ser-239 by protein kinase A: Control of subcellular localization. J Lipid Res. 2014 doi: 10.1194/jlr.M055004. jlr. M055004. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten A, Seline A, Rudd A, Logsdon M, Listenberger LL. Surface features of the lipid droplet mediate perilipin 2 localization. Biochem Biophys Res Commun. 2014;452:422–427. doi: 10.1016/j.bbrc.2014.08.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak JC, Hayden CC, Sasaki DY. Steric confinement of proteins on lipid membranes can drive curvature and tubulation. Proc Natl Acad Sci U S A. 2010;107:7781–7786. doi: 10.1073/pnas.0913306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, Hayden CC. Membrane bending by protein-protein crowding. Nat Cell Biol. 2012;14:944–949. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- Stone SJ, Levin MC, Farese RV., Jr Membrane topology and identification of key functional amino acid residues of murine acyl-CoA:diacylglycerol acyltransferase-2. J Biol Chem. 2006;281:40273–40282. doi: 10.1074/jbc.M607986200. [DOI] [PubMed] [Google Scholar]
- Thiam AR, Antonny B, Wang J, Delacotte J, Wilfling F, Walther TC, Beck R, Rothman JE, Pincet F. COPI buds 60-nm lipid droplets from reconstituted water-phospholipid-triacylglyceride interfaces, suggesting a tension clamp function. Proceedings of the National Academy of Sciences of the United States of America. 2013a;110:13244–13249. doi: 10.1073/pnas.1307685110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam AR, Farese RV, Jr, Walther TC. The biophysics and cell biology of lipid droplets. Nature reviews Molecular cell biology. 2013b;14:775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel K, Heier C, Haberl V, Thul PJ, Oberer M, Lass A, Jäckle H, Beller M. The evolutionarily conserved protein CG9186 is associated with lipid droplets, required for their positioning and for fat storage. J Cell Science. 2013;126:2198–2212. doi: 10.1242/jcs.120493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annual review of biochemistry. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Martin DD, Genter E, Wang J, McLeod RS, Small DM. Surface study of apoB1694-1880, a sequence that can anchor apoB to lipoproteins and make it nonexchangeable. J Lipid Res. 2009;50:1340–1352. doi: 10.1194/jlr.M900040-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JJ, Wong KF, Ansell SM, Masin D, Bally MB. Polyethylene glycol modified phospholipids stabilize emulsions prepared from triacylglycerol. J Pharm Sci. 1994;83:1558–1564. doi: 10.1002/jps.2600831108. [DOI] [PubMed] [Google Scholar]
- Wilfling F, Thiam AR, Olarte MJ, Wang J, Beck R, Gould TJ, Allgeyer ES, Pincet F, Bewersdorf J, Farese RV, Jr, Walther T. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife. 2014;3 doi: 10.7554/eLife.01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Developmental Cell. 2013;24:384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolins NE, Brasaemle DL, Bickel PE. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006;580:5484–5491. doi: 10.1016/j.febslet.2006.08.040. [DOI] [PubMed] [Google Scholar]
- Wolins NE, Rubin B, Brasaemle DL. TIP47 associates with lipid droplets. J Biol Chem. 2001;276:5101–5108. doi: 10.1074/jbc.M006775200. [DOI] [PubMed] [Google Scholar]
- Xie X, Langlais P, Zhang X, Heckmann BL, Saarinen AM, Mandarino LJ, Liu J. Identification of a novel phosphorylation site in adipose triglyceride lipase as a regulator of lipid droplet localization. 2014 doi: 10.1152/ajpendo.00663.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanghellini J, Natter K, Jungreuthmayer C, Thalhammer A, Kurat CF, Gogg-Fassolter G, Kohlwein SD, von Grünberg HH. Quantitative modeling of triacylglycerol homeostasis in yeast--metabolic requirement for lipolysis to promote membrane lipid synthesis and cellular growth. FEBS J. 2008;275:5552–5563. doi: 10.1111/j.1742-4658.2008.06681.x. [DOI] [PubMed] [Google Scholar]
- Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res. 2009;50:3–21. doi: 10.1194/jlr.R800031-JLR200. [DOI] [PubMed] [Google Scholar]
- Zehmer JK, Bartz R, Liu P, Anderson RG. Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets. J Cell Sci. 2008;12:1852–1860. doi: 10.1242/jcs.012013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman SB, Minton AP. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.