Abstract

The development of methods for achieving precise spatiotemporal control over chemical and biomolecular gradients could enable significant advances in areas such as synthetic tissue engineering, biotic–abiotic interfaces, and bionanotechnology. Living organisms guide tissue development through highly orchestrated gradients of biomolecules that direct cell growth, migration, and differentiation. While numerous methods have been developed to manipulate and implement biomolecular gradients, integrating gradients into multiplexed, three-dimensional (3D) matrices remains a critical challenge. Here we present a method to 3D print stimuli-responsive core/shell capsules for programmable release of multiplexed gradients within hydrogel matrices. These capsules are composed of an aqueous core, which can be formulated to maintain the activity of payload biomolecules, and a poly(lactic-co-glycolic) acid (PLGA, an FDA approved polymer) shell. Importantly, the shell can be loaded with plasmonic gold nanorods (AuNRs), which permits selective rupturing of the capsule when irradiated with a laser wavelength specifically determined by the lengths of the nanorods. This precise control over space, time, and selectivity allows for the ability to pattern 2D and 3D multiplexed arrays of enzyme-loaded capsules along with tunable laser-triggered rupture and release of active enzymes into a hydrogel ambient. The advantages of this 3D printing-based method include (1) highly monodisperse capsules, (2) efficient encapsulation of biomolecular payloads, (3) precise spatial patterning of capsule arrays, (4) “on the fly” programmable reconfiguration of gradients, and (5) versatility for incorporation in hierarchical architectures. Indeed, 3D printing of programmable release capsules may represent a powerful new tool to enable spatiotemporal control over biomolecular gradients.
Keywords: 3D printing, spatiotemporal patterning, release capsules, plasmonic nanorods, core–shell particles, biomolecular gradients
The ability to mimic the dynamic microenvironment surrounding cells in natural tissues is critical to engineering synthetic analogues.1–5 Cell fate is influenced by numerous molecular factors and interactions that require meticulous control for regeneration of functional tissue. In order to achieve such control, engineered matrices must be capable of generating multiplexed spatiotemporal molecular gradients in 3D architectures.6–8 Extensive research efforts to engineer such matrices have resulted in a number of promising methods to generate and control molecular gradients.1,9–14 For example, novel labile chemical linkers have been used to pattern and release biomolecules from polymer backbones in response to external stimuli.13, 15 While this method provides excellent spatiotemporal control, selective multiplexed release requires engineering orthogonal linkers for each molecular factor to be released, which can quickly become cumbersome. As a second example, microfluidic channels have been directly incorporated in hydrogels, providing a means to flow biomolecule solutions through the gels.12, 16, 17 This allows for the flexibility of generating steady-state gradients that can be maintained over long periods of time.12 However, multiple independent networks are required for multiplexed gradients, and an extensive pumping and fluid-handling infrastructure is required. This approach is thus more suitable as a route to generate vasculature by consistently supplying nutrients to and removing waste from the tissue, rather than a means of generating transient gradients of biomolecules that can control cell fate at a local level.18
Micro/nanoparticles loaded with biomolecules represent a versatile approach to delivering multiplexed gradients.6,19–22 While such particles can be made via numerous methods and from a wide range of materials, they are most commonly formulated from biodegradable polymers using double emulsification or coacervation methods.23 The particles can be efficiently loaded with a variety of biomolecular payloads, while maintaining their activities. Synthesizing particles loaded with different factors and localizing them within a hydrogel matrix can lead to the generation of multiplexed spatial gradients.6 The payload release kinetics can be adjusted by controlling particle properties (e.g., diameter, shell thickness, and porosity). However, spatiotemporal control over the gradients is typically coarse, as most scalable methods to synthesize particles result in highly polydisperse populations.24 Recent research efforts have focused on two significant challenges to achieving robust control over release kinetics. First, microfluidic methods are being developed to synthesize monodisperse particles with a high degree of control over particle properties.25–27 Second, strategies to render particles stimuli-responsive are being developed, so that the release of biomolecules can be manipulated externally and dynamically.20, 28–31
3D printing is an additive manufacturing technique with the potential to revolutionize the fabrication of bioactive nanodevices containing biotic–abiotic interfaces, for applications such as tissue engineering scaffolds.32–36 In prior work, we have shown that 3D printing offers the flexibility to interweave a wide palette of functional materials, including nanomaterials, biomaterials, and living cells, in a free-form, layer-by-layer manner.32, 37–39 Moreover, in biomedical applications, each device can be customized to patient-specific designs and functionalities. Finally, the recent explosion of interest in this table-top manufacturing technology has dramatically lowered the technical barriers for entry and use.
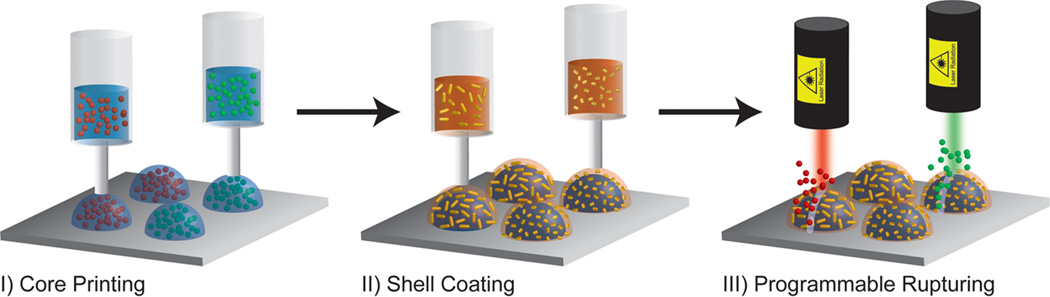
Here, we introduce a novel 3D printing based method to produce highly monodisperse core/shell capsules with robust control over particle properties, passive release kinetics, and particle distributions throughout a 3D matrix. Furthermore, we render these capsules stimuli-responsive by incorporating AuNRs into the polymer shell, allowing for highly selective photothermal rupture and triggered temporal release of the biomolecular payload. Figure 1 illustrates our concept. First, a patterned array of multiple aqueous cores containing functional biomolecules is printed on a solid substrate. Second, polymer shells functionalized with AuNRs are deposited to encapsulate each printed core. All of the fabrication processes are accomplished using a custom-built 3D printer (see Supporting Information) based on a fluid-dispensing robot combined with a digital pressure regulator. Finally, the programmable release of functional biomolecules is triggered by laser rupture of the polymer shell due to the photothermal effect caused by the localized surface plasmon resonance (LSPR) of the AuNRs.40
Figure 1.
Programmable printing and rupturing of capsules: (I) multiplexed arrays of aqueous cores containing biomolecular payloads are printed directly on a solid substrate; (II) PLGA solutions containing AuNRs of varying lengths are dispensed directly on the aqueous cores, forming a solid stimuli-responsive shell; (III) the capsules are selectively ruptured via irradiation with a laser wavelength corresponding to the absorption peak of the nanorods.
A critical first step is to develop reliable methods to print the aqueous cores, which contain the target biomolecules, with defined control over volumes and concentrations. Cores were printed using a 3D printer based on a Fisnar F5200N benchtop gantry (see Supporting Information). To tune the printability of the aqueous core inks, poly(vinyl alcohol) (PVA, MW ~ 85 000; Sigma-Aldrich) was added to adjust the ink to a suitable viscosity, and ethylene glycol (J.T. Baker) was added as a humectant. A typical core ink formulation consisted of 0.5 g of PVA and 2.5 g of ethylene glycol dissolved in 10 g of total solution. Into this solution, payloads such as food dyes, biomolecules, or enzymes were dissolved at concentrations ranging from 0.1 to 10 mg/mL.
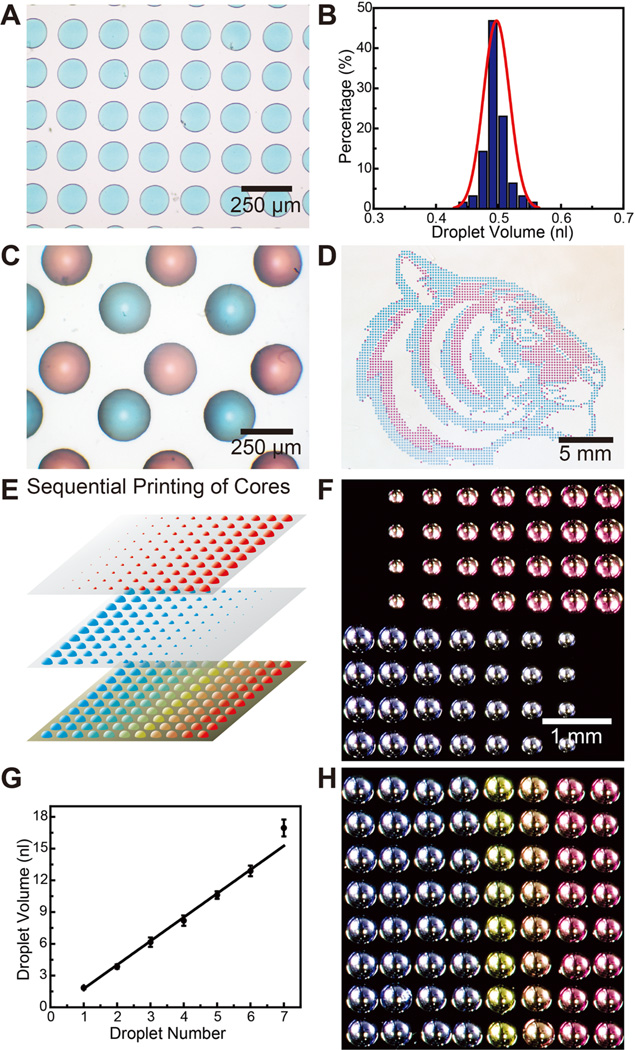
Aqueous cores were first printed onto a hydrophobic substrate such as a polystyrene-coated Si wafer or siliconized glass slides (Hampton Research). Moderately hydrophobic substrates were selected (contact angles ~ 70°–100°) so that aqueous cores could partially wet the substrate and detach from the printer tip, while minimizing spreading and maintaining the droplet shape. The volume of dispensed droplets was controlled by varying the dispense time, applied pressure, and distance between tip and substrate. Figure 2A shows a representative region of a 25 × 25 array of blue aqueous cores with a 200 µm center-to-center spacing printed on a polystyrene substrate. The diameters of the printed cores can be controlled via proper modulation of the dispensing time and pressure during printing. As shown in Figure 2B, the mean core volume is 0.5 nL, with a total volume variation of less than 10%. To demonstrate spatial control, Figure 2C shows an optical image of an array of red and blue multiplexed cores with center-to-center spacings of 200 µm between red and blue neighbors. Significantly, the patterned arrays can be produced over large areas with high fidelity. Figure 2D shows the printing of a “Princeton tiger” consisting of 4000 red and blue cores, with a spacing of 400 µm.
Figure 2.
Printing of aqueous cores. (A) Optical micrograph of an array of blue aqueous cores with a center-to-center spacing of 200 µm (colors are from commercial food dyes). (B) The corresponding histogram shows the distribution of droplet volumes. (C) Optical micrograph of a multiplexed array of red and blue capsule cores with spacing of 200 µm between cores of opposite color. (D) Optical image of large scale (>4000) multiplexed patterned red and blue capsule cores in the shape of a “Princeton tiger”. (E) Schematic illustrating the generation of capsule arrays with varying compositions. Arrays with varying volumes are generated by dispensing multiple droplets in the same locations. Overlaying them creates arrays with varying compositions. (F) Optical micrographs of two different pH capsule arrays with varying core volumes. The number of dispensed drops varies from 1 to 8 drops across the array. (G) Plot showing a linear relationship of core volume to the number of droplets dispensed. (H) Optical micrograph of a 2D array with varying pH, adjusted by the ratio of H2SO4 and NaOH within, indicated by the color of m-cresol purple.
To demonstrate the versatility and 2D multiplexing capabilities of this approach, we next sought to generate arrays of cores containing varying volumes and compositions,41 as illustrated in Figure 2E. The ability to dispense finely tuned volumes allowed us to adjust the core volumes by simply printing multiple droplets into the same core. Figure 2F shows two printed core arrays with the volume gradient variations mapped according to Figure 2E. The different colors between the two arrays were due to the different pH values of the cores, which was adjusted by varying the ratios of H2SO4 and NaOH inside, as indicated by the color of added m-cresol purple (red: pH = 1.25; violet: pH = 11.90; Sigma-Aldrich). A linear relationship between the core volume and the number of printed droplets was observed, as shown in Figure 2G. Next, we created capsule arrays with compositional gradients by overlaying multiple arrays of varying volumes. Specifically, we created an array of cores with varying pH values via the chemical reaction of the two printed cores. The final array with varying colors is shown in Figure 2H, suggesting that the pH values of the cores became more acidic from left to right (pH values: 11.90, 11.70, 11.42, 10.74, 3.59, 1.64, and 1.25). This ability to readily generate arrays with a multifactorial composition gradient could be useful for combinatorial screening applications.42, 43
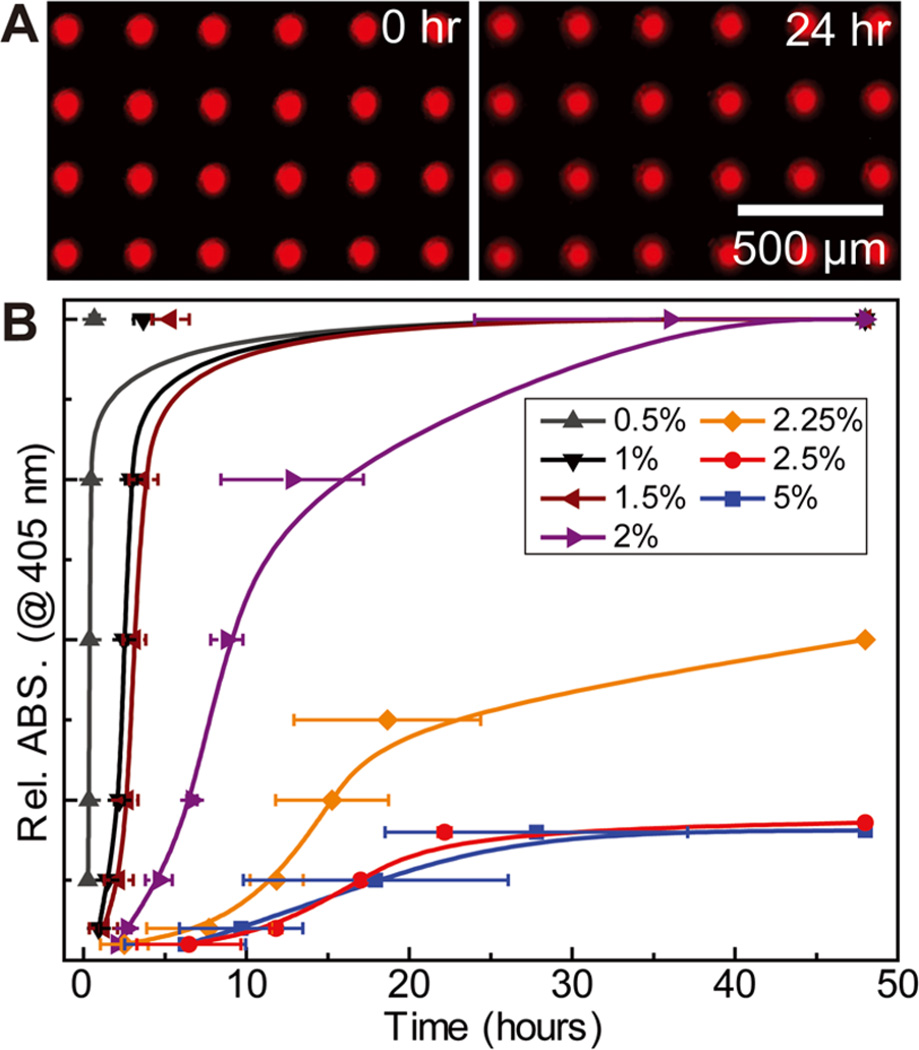
Next, shells were dispensed onto the capsule cores. In order to encapsulate the aqueous cores and achieve controllable core release, we developed a method to dispense a PLGA solution directly on the aqueous core, which dried into a uniform shell. Shell inks were prepared with PLGA (75:25, MW ~ 65 000– 107 000; Sigma-Aldrich) concentrations ranging from 0.5 to 5 wt % in dichloromethane (Sigma-Aldrich). Figure 3A shows the encapsulation of an aqueous core containing fluorescent dextran (40 kDa) by a shell formed from 2.5 wt % PLGA solution. The capsules were immersed in water to determine the leakage of dextran molecules through any potential defects in the polymer shell. The fluorescent images show the array at 0 and 24 h, suggesting minimal burst release and hence minimal shell defects. This is in contrast to typical solution-based preparations of polymer microcapsules, which yield capsules that have a well-known burst release over the first 24 h, caused by porous defects in the polymer shells.44 A key feature in this case that allows the PLGA solution to form uniform shells is that while the polymer solution and aqueous droplet are immiscible, the PVA in the core acts as an emulsifier and allows the polymer solution to fully wet the core droplet. Additionally, the high volatility and small volume allow the shell to rapidly solidify, forming a kinetically stabilized shell structure.
Figure 3.
Encapsulation of the aqueous core with a PLGA shell. (A) Fluorescent optical micrographs showing the stability of encapsulated cores containing Rhodamine B isothiocyanate–dextran submerged in water over a 24 h period. (B) Plots showing tunable passive release kinetics of HRP from capsule arrays, monitored by an ABTS substrate, achieved by varying the thickness of the PLGA shell (N = 3).
We next investigated whether adjusting the shell thickness by varying the PLGA concentration in the ink could provide control over the passive diffusion and release of biomolecules from the capsule cores. To determine this, we encapsulated horseradish peroxidase (HRP, Type I, 52 units/mg, Sigma-Aldrich) in capsules with PLGA shells of varying thicknesses. Figure 3B shows the passive enzyme release, monitored by the colorimetric reaction of one-step 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS, Thermo Scientific) over 48 h. The release of fluorescent dyes was directly observed by fluorescent microscopy. To perform the assay, 10 × 10 arrays containing 1 mg/mL HRP were encapsulated with PLGA shells ranging from 0.5 to 5 wt %. A constant volume (50 µL) of PLGA solution was used for the encapsulation to ensure minimal shell thickness variations. The arrays were submerged in 5 mL of a reaction medium composed of 1 mL one-step ABTS solution and 4 mL water in a six-well plate. The reaction was monitored by sampling 100 µL of the reaction medium and measuring the absorbance at 405 nm using a spectrophotometer over a 48 h time period. It can be seen that with increasing PLGA concentrations the HRP release rate decreased gradually, demonstrating that the passive molecular release can be tailored over a broad release profile by simply varying the polymer concentration in the shell ink. This is in contrast to solution-based methods, where release profiles depend on several processing variables and require a greater degree of optimization to achieve such control.23, 24 When the shell was printed from higher concentration PLGA solutions (≥2.5 wt %), the cores were well-encapsulated with minimal passive release. Thus, these high concentration PLGA films hold the highest potential as shell materials for stimuli-responsive release studies.
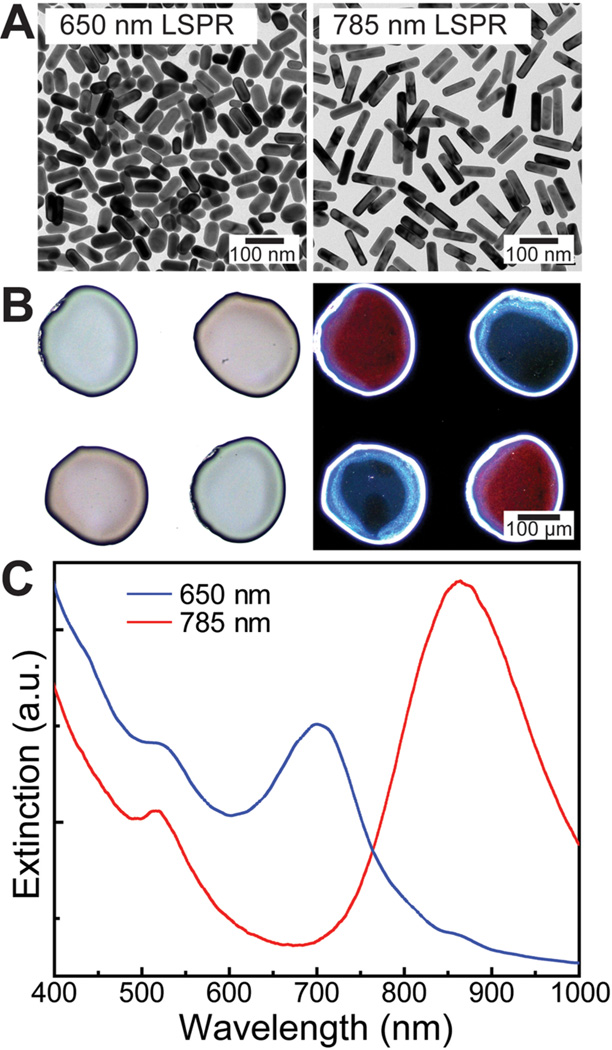
To achieve programmable control over 3D gradients, we incorporated AuNRs in the capsule shells. AuNRs strongly absorb and scatter light at the LSPR wavelength, which is dependent on the aspect ratio of the nanorod.45, 46 This absorption is nonradiative; hence, the absorbed energy is converted to heat, which suggests that nanorods are promising candidates for imparting a photothermal response.47 Here, we utilized the nanorods to locally heat and rupture the capsule shells. The sharp and tunable absorption makes them ideal to provide highly selective stimuli-responsive rupture to the multiplexed arrays. We utilized two different length nanorods (diameter 25 nm, Nanopartz Inc.) with vis–NIR absorption peaks at 650 and 785 nm at 100 OD/mL (Figure 4A). In order to incorporate the nanorods in the polymer shell, we utilized nanorods functionalized with a polymeric coating, which allowed the nanorods to be easily dispersed in the polymer solution (see Supporting Information). To prepare the ink, the AuNR solution was mixed with an equal part of 5 wt % PLGA in dichloromethane, yielding a solution with a final concentration of 2.5 wt % PLGA and 2.5 OD/mL AuNRs in dichloromethane.
Figure 4.
Incorporation of AuNRs in the PLGA shell. (A) TEM images of two different length AuNRs (diameter ~25 nm) with absorption peaks centered at 650 and 785 nm. (B) Bright-field (left) and dark-field (right) optical micrographs of dispensed PLGA shell arrays functionalized with different length nanorods. In the bright-field image, the blue and red colors correspond to the 650 and 780 nm absorption nanorods. The colors are switched in the dark-field image because this shows the scattered component of light as compared to the transmitted in bright-field mode. (C) Vis–NIR spectra of PLGA shells loaded with different length nanorods.
Figure 4B shows optical bright-field and dark-field images (transmitted and scattered components of light, respectively) of polymer shells (2.5 wt % PLGA) incorporating both lengths of nanorods printed in an alternating fashion. The observed colors of the printed shells indicate the successful incorporation of nanorods within the PLGA polymer, while the high contrast of the two kinds of nanorod-functionalized shells suggests the promising application of selective optical rupture. To confirm the LSPR wavelengths of the immobilized AuNRs for laser selection in photothermal release, the vis–NIR spectra of the printed capsule shells were measured as shown in Figure 4C. The spectra appear red-shifted (650 → 695 nm, 785 → 855 nm) due to the higher refractive index of the polymer compared to aqueous solutions. Minimizing aggregation of the nanorods is essential to maintaining the sharpness and location of absorption that allows for the selective multiplexed stimuli response. The lack of shoulder peaks or a large shift in the plasmon peaks indicates that nanorods were indeed well dispersed in the PLGA shells.45
To test the ability to selectively rupture and release biomolecular payloads at different wavelengths, we encapsulated HRP-containing cores with capsule shells containing either 650 or 785 nm LSPR nanorods. PLGA/AuNR shells were dispensed directly over the capsule array with a controlled volume (50 µL/cm2). The encapsulated array was then either submerged in aqueous media or covered with a layer of hydrogel. As discussed previously, PLGA solutions of higher concentrations were ideal for forming robust capsule shells for photothermal rupture. Comparing the data of 2.5 and 5.0 wt % PLGA in passive release experiments (red and blue curves in Figure 3b, respectively), it can be observed that the 2.5 wt % PLGA shell kept the enzyme encapsulated with slightly higher efficiency as compared to the 5 wt % shell. This may be due to the potential for greater film defect formation in the higher concentration solutions caused by irregular drying. Additionally, the lower concentration polymer solution produces a thinner capsule shell, which is more favorable for photothermal rupture. Therefore, we selected 2.5 wt % PLGA as the optimized capsule shell for laser-triggered molecular release.
Diode lasers (Power Technology, Inc.) with emissions at 658 and 783 nm and intensities of 100 and 125 mW, respectively, were used for the rupturing measurements. The laser spot size was focused to approximately 30 µm, so that capsules could be individually addressed with high resolution (see Supporting Information). In order to accurately position the lasers, they were directly mounted on the benchtop gantry arm using a custom fabricated holder. The laser rupture pathway was programmed using the Smart Robot software with line speeds ranging from 5 to 10 mm/s. The optimal laser intensity and concentration of nanorods in the PLGA films were determined in both air and water ambient conditions (see Supporting Information). The laser rupture and functional molecule release was first demonstrated by an HRP solution covered by a bulk PLGA film. As shown in Movie 1, the 650 nm nanorod-functionalized PLGA film was ruptured via parallel passes by a laser of wavelength 658 nm, and the solution containing the HRP enzymes was gradually released. To measure laser-triggered HRP release from a single capsule, the fluorometric reaction with the Amplex Red (Life Technologies) reagent was monitored using fluorescence microscopy. The HRP encapsulated array was covered with a hydrogel ambient containing the Amplex Red reagent. The hydrogel was prepared by adding 1 mL of 5× reaction buffer to 4 mL of 40 wt % Pluronic F-127 (Sigma-Aldrich) aqueous solution at 4 °C. To this solution, 50 µL of a 10 mM Amplex Red solution in DMSO and 25 µL of 3% H2O2 were added just prior to use. The assay was performed by dispensing approximately 100 µL of hydrogel medium on a HRP capsule array.
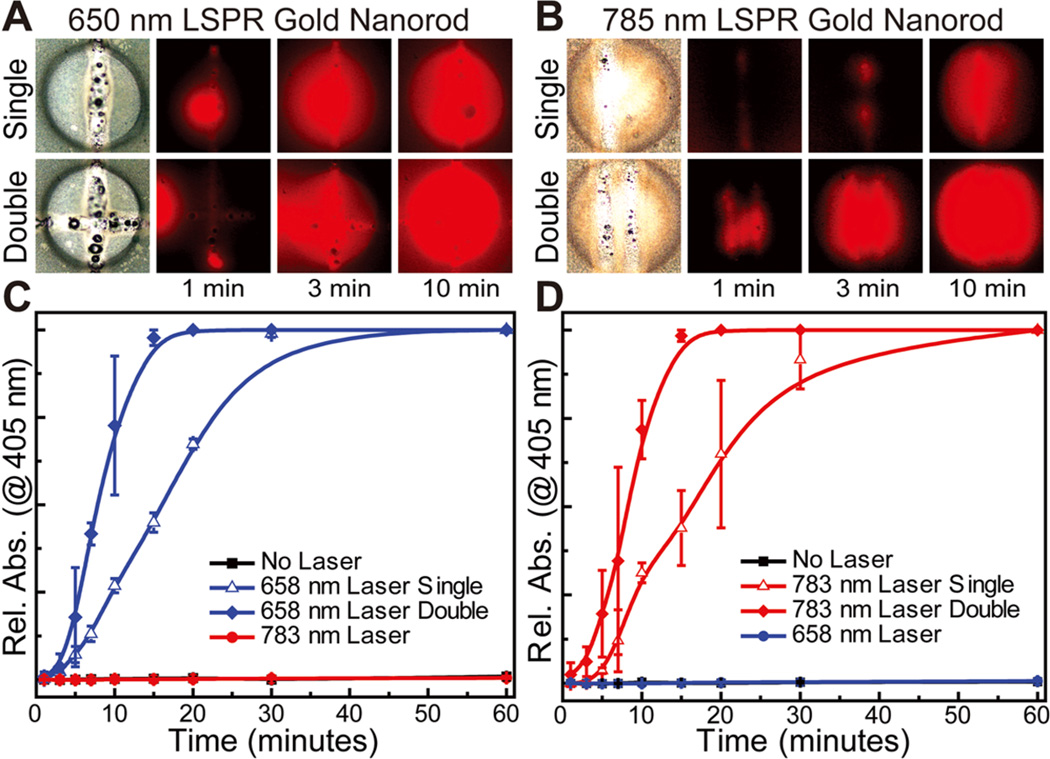
Figures 5A and 5B show optical bright-field and fluorescent images of ruptured capsules with 650 and 785 nm nanorod-modified shells, respectively. The top image row shows capsules that have been ruptured with a single pass of the laser, and the bottom row shows capsules that have been ruptured with a double pass. The fluorescent images were taken at 1, 3, and 10 min after laser irradiation and show the progression of the reaction between the released HRP and fluorometric substrate (Amplex Red) present in the ambient hydrogel. Next, we quantitatively analyzed these selective biomolecular releases over an entire array of capsules that had been ruptured with a single pass, double pass, or exposed to the opposite laser, by monitoring the colorimetric reaction of ABTS in 1 h. These experiments are shown in Figures 5C and 5D for 650 and 785 nm nanorod-modified shells, respectively.
Figure 5.
Selective laser-triggered rupture and release of enzymes. (A, B) Bright-field and fluorescent optical images of 650 and 785 nm ruptured capsules showing release of HRP (monitored by Amplex Red), respectively, with release rate controlled by the ruptured area of the PLGA polymer shell (edge length of the square images: 300 µm) (C, D) Plots showing release of HRP from ruptured capsule arrays, demonstrating orthogonal selectivity of AuNR-encrusted capsules as well as fine control over release kinetics, monitored by an ABTS substrate (N = 3).
Significantly, both the visible optical images and quantitative absorption plots clearly demonstrate that the capsules are ruptured only when exposed to their corresponding laser wavelengths, and thus the stimuli response is orthogonal and selective. Moreover, we showed that by manipulating the number of passes of the laser over the ruptured area of the capsule, the release rate of the payload could be very accurately controlled even at very short time scales. This capability provides control over the temporal release of biomolecules within the capsule array even at the level of a single capsule. This high-resolution capability to address individual capsules can further be extended over an entire array to rupture capsules in a programmable manner (see Supporting Information). The critical role of AuNRs in the photothermal rupture of the PLGA shells was further verified by control experiments showing that nonfunctionalized shells remained pristine when exposed to both laser sources. Additionally, we observed that efficient heating and rupturing of the shells was also dependent on nanorod diameter. Specifically, the PLGA polymer shell could not be fully ruptured if smaller diameter nanorods (10 nm) were incorporated, instead of the larger 25 nm diameter nanorods. This finding is expected, as the extinction coefficient increases with the nanorod diameter.48
The localized heating due to absorption in the AuNRs is expected to minimally impact the surrounding microenvironment. However, since the microenvironment can host biologically active species whose function is degraded at elevated temperatures—for example, biomolecules within the core and cells within the surrounding matrix—we examined the associated heat transfer characteristics based on the experimental conditions (see Supporting Information). The laser spot size (DL ~ 30 µm) and the experimentally observed rupture area (ca. 40 µm, from Figure 5) are much smaller than the particle diameter (Dp ~ 200 µm), and the polymer shell thickness (~10 µm) is significantly less than both Dp and the hydrogel thickness (tg > 1 mm). Therefore, it is useful to model the associated thermal effects as a one-dimensional conduction problem with a boundary heat source and a semi-infinite domain. Given that the dissipation of heat by absorbed and scattered photons, and the conduction of heat in the nanorod-functionalized shells, are much more rapid than the conduction within the inner aqueous core and the surrounding hydrogel matrix, the resultant temperature profile within the hydrogel is described by49
| (1) |
Here, T(x,t) is the temperature in the hydrogel at position x and time t, Ts is the steady state temperature of the nanorodfunctionalized polymer shell, Ti is the initial temperature of the hydrogel (room temperature), and α = k/ρCp is the thermal diffusivity, where k, ρ, and Cp are the hydrogel thermal conductivity, density, and heat capacity, respectively.
We assume that α is similar to the thermal diffusivity of water, 0.143 × 10−6 m2/s;50 Ti = 22 °C, and since the melting temperature of the polymer is 40–60 °C, we used Ts = 50 °C. To determine the heating impact on the microenvironment, it is useful to calculate the heat penetration depth (tpen) into the surrounding hydrogel, as excessive heating may cause cell damage. Considering that the experimentally observed rupture was approximately 0.03 s (i.e., LD/laser scan rate of 1 mm/s), and assuming that loss of biological activity in cells occurs when the temperature exceeds 40 °C, only the immediately surrounding hydrogel matrix within tpen = 43 µm of the nanorod-functionalized shell would reach the threshold temperature. The remainder of the hydrogel would remain below the threshold temperature. On the other hand, the threshold temperature will vary inside the core. The aqueous core is expected to have a higher tolerance, as biomolecules are typically more robust than cells. Here, we employed HRP as the model in the selective release experiment. Although enzymes exhibit lower thermal stability compared to other biomolecules, such as DNA and small molecular drugs, the denaturation temperature of the enzyme is ~70 °C (the threshold temperature of the core). This is sufficiently high that the brief exposure to the laser light is highly unlikely to lead to denaturation.
The approach described provides an excellent means of generating 2D arrays of capsules on a solid substrate, for selectively programmable biomolecular release. These arrays have tremendous potential as a spatiotemporal platform to controllably probe the effects of multiplexed biomolecular gradients. A significant challenge is to create 3D arrays of the capsules,51 but this requires that the aqueous core be fully encapsulated without an underlying substrate. To address this, we developed a new type of ink based on a water-in-oil emulsion of the aqueous core in the PLGA solution (Figure 6A). The emulsion inks were prepared via high-speed dispersion of aqueous core solutions into the PLGA/AuNR solution. The core was an aqueous solution of food or fluorescent dyes—green (poly(fluorescein isothiocyanate allylamine hydrochloride), Sigma-Aldrich) or red (Rhodamine B isothiocyanate-dextran (MW ~ 40 000, Sigma-Aldrich))—at concentrations ranging from 0.1 to 1 mg/mL. The PLGA/AuNR solution was prepared with 10 wt % PLGA and 2.5 OD/mL AuNRs (780 nm absorption) in dichloromethane. To prepare the emulsion, 200 µL of the aqueous core was added to 800 µL of the PLGA/AuNR solution and dispersed using an IKA T10 disperser at 30 000 rpm for 60 s. Care was taken to adjust the viscosity and density of both the aqueous and organic phases to limit separation of the dispersed droplets during the printing process. In order to avoid the direct passive release of the aqueous core, a 10 wt % PLGA solution was employed to form the polymer shell in this emulsion-type ink.
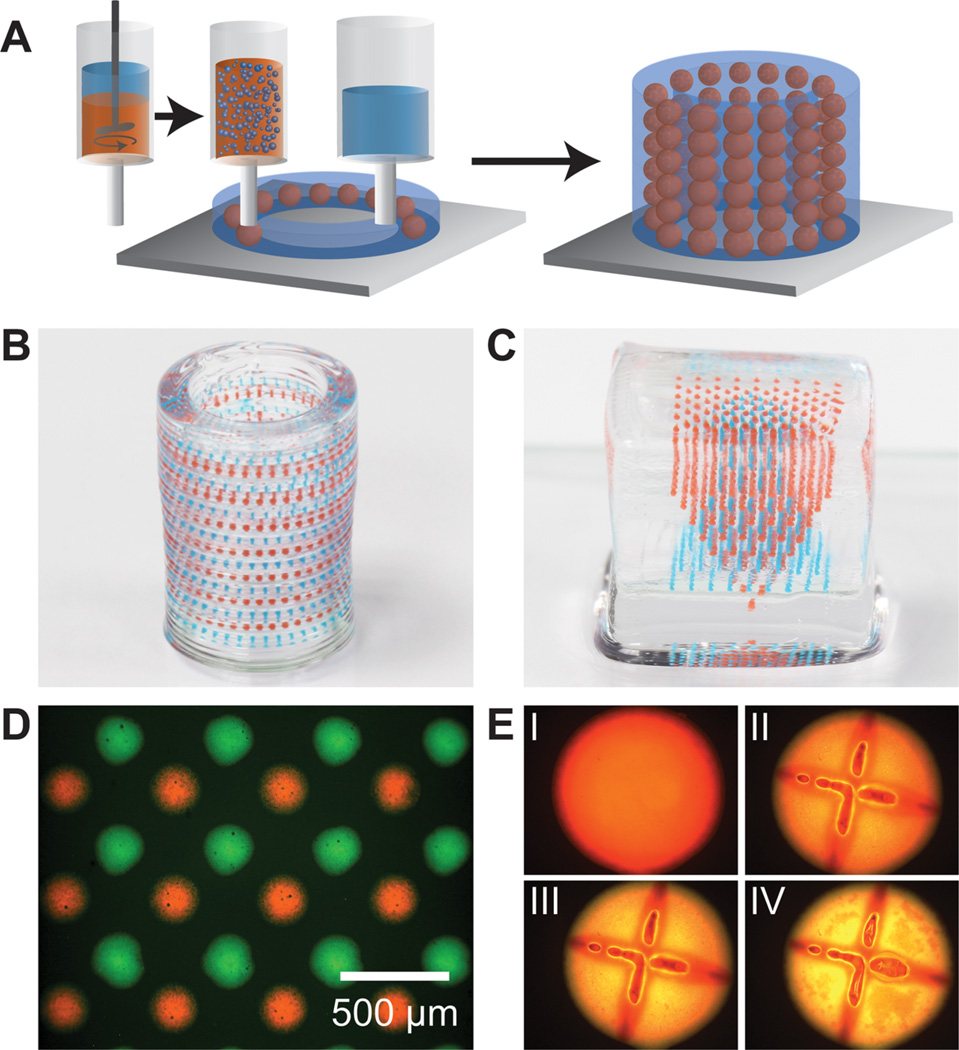
Figure 6.
3D printing of hierarchically multiplexed capsule arrays. (A) Schematic illustrating an emulsion ink-based method to 3D print complex capsule arrays. The emulsion ink is prepared by directly dispersing the aqueous core in the PLGA solution. The hydrogel and emulsion inks are sequentially printed in a layer-by-layer manner to form a 3D structure. (B, C) Optical images of 3D multiplexed capsule arrays directly printed in cylindrical and square hydrogel matrices, respectively (colors of the capsules are from food dyes in the dispersed cores). (D) Fluorescent optical image of a single layer of a multiplexed emulsion-based capsule array. (E) Fluorescent optical images showing rupture and release of fluorescein dye (poly(fluorescein isothiocyanate allylamine hydrochloride)) from an emulsion capsule with Nile red stained PLGA (I: before laser rupture; II, III, IV: 15 min, 1 h, and 2 h after laser rupture; diameter of the capsule: ~300 µm).
As shown in the schematic, the emulsion-based ink was directly printed into a thin layer of an aqueous hydrogel. Once printed, the solvent rapidly evaporated through this layer, leaving behind a solidified capsule. Thus, the hydrogel and capsules can be readily printed in a layer-by-layer fashion to create complex 3D hierarchical programmable capsule arrays. A wide range of hydrogels can be easily incorporated. For these experiments, we utilized a hydrogel based on Pluronic F-127 that gels at concentrations above 20 wt % at room temperature. The hydrogel ink was prepared using a 40 wt % solution of Pluronic F-127 in water. 3D arrays were printed sequentially by depositing the hydrogel and emulsion inks in a layer-by-layer manner (Movie 2). First, the base layers of the hydrogel ink were dispensed on a glass slide. Next, the emulsion ink was directly dispensed into the hydrogel layer. This sequence was repeated to create free-form hierarchical structures with multiple capsule layers. The structure of the hydrogel medium and the patterns of emulsion capsules were programmed with the Smart Robot software.
Since the 3D printing process is based on digital software control, any number of rationally designed advanced architectures can be constructed by this method. For instance, Figure 6B shows an optical photograph of a hollow hydrogel cylinder containing alternating layers of red and blue capsules in the cylinder wall. The process for 3D printing the structure is shown in Movie 2. Another example of a complex 3D structure is shown in Figure 6C, consisting of two inverted pyramidal arrays of capsules printed within a solid hydrogel cube. Precise control over spatial patterning can be thus demonstrated by this emulsion ink-based printing.
The fluorescent microscope image in Figure 6D shows a single layer of a multiplexed capsule array containing fluorescently tagged dextrans in dispersed aqueous droplets, of which the resolution (center-to-center spacing of ~200 µm between red and green neighbors) is similar to the capsule arrays fabricated by sequential printing. Furthermore, we demonstrated the stimuli-responsiveness of these capsules by releasing water-soluble fluorescein dye. Figure 6E (panel I) shows a printed capsule with a fluorescein (poly(fluorescein isothiocyanate allylamine hydrochloride)) tagged aqueous core and Nile red tagged PLGA shell functionalized by AuNRs. Once ruptured (panels II through IV) by the corresponding laser, the release of fluorescein can be observed via the increasing green/yellow color. Thus, these emulsion-based inks fully enable complex 3D arrays with selective spatiotemporal response and functionality.
In summary, we have developed a novel technique to directly print selectively programmable release capsules in multiplexed arrays. This demonstration represents a proof of concept to illustrate that the combination of additive manufacturing techniques and functional plasmonic nanomaterials can facilitate the hierarchical assembly of materials in new and unique ways. Here, 3D printing allowed us to efficiently generate highly monodisperse capsules in well-defined spatial patterns, while being able to independently and precisely control the core volumes, compositions, and shell thicknesses. The addition of AuNRs imparted a highly selective stimuli response, allowing for the rupture and release of biomolecules from these multiplexed arrays. Finally, implementing an emulsion-based ink allowed us to extend this approach to complex 3D arrays directly interwoven within a hydrogel matrix. Future studies will focus on several remaining challenges, including (1) improving the resolution and spatial alignment of these printed capsules, (2) developing stimuli-responsive shells with reversible payload release properties, (3) developing methods to quantify in real time the concentrations of payload molecules within the capsule cores, and (4) proving the biocompatibility and applicability of 3D matrices with printed capsules at the cellular level.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Rajesh R. Naik, Howard A. Stone, and Nan Yao for useful discussions and advice. The authors thank Jesse Goodman and Huai-An Chin for their technical help. We acknowledge the use of the PRISM Imaging and Analysis Centre, which is supported by the NSF MRSEC Program via the Princeton Centre for Complex Materials (No. DMR-0819860). M.K.G. was supported by an Intelligence Community Postdoctoral Fellowship (Award 2013-13070300004). M.C.M. and S.S. acknowledge support of this work by the Air Force Office of Scientific Research (Award FA9550-12-10368) and M.C.M. acknowledges support by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health (Award 1DP2EB020537). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
ASSOCIATED CONTENT
Additional experimental details with materials, methods, and figures. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.5b01688.
The authors declare no competing financial interest.
REFERENCES
- 1.Biondi M, Ungaro F, Quaglia F, Netti PA. Adv. Drug Delivery Rev. 2008;60:229–242. doi: 10.1016/j.addr.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 2.Griffith LG, Swartz MA. Nat. Rev. Mol. Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 3.Lutolf MP, Gilbert PM, Blau HM. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lutolf MP, Hubbell JA. Nat. Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 5.Kalinin YV, Murali A, Gracias DH. RSC Adv. 2012;2:9707–9726. doi: 10.1039/C2RA20337E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen FM, Zhang M, Wu ZF. Biomaterials. 2010;31:6279–6308. doi: 10.1016/j.biomaterials.2010.04.053. [DOI] [PubMed] [Google Scholar]
- 7.Cao L, Mooney DJ. Adv. Drug Delivery Rev. 2007;59:1340–1350. doi: 10.1016/j.addr.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keenan TM, Folch A. Lab Chip. 2008;8:34–57. doi: 10.1039/b711887b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Causa F, Netti PA, Ambrosio L. Biomaterials. 2007;28:5093–5099. doi: 10.1016/j.biomaterials.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 10.Khademhosseini A, Langer R. Biomaterials. 2007;28:5087–5092. doi: 10.1016/j.biomaterials.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 11.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi NW, Cabodi M, Held B, Gleghorn JP, Bonassar LJ, Stroock AD. Nat. Mater. 2007;6:908–915. doi: 10.1038/nmat2022. [DOI] [PubMed] [Google Scholar]
- 13.Luo Y, Shoichet MS. Nat. Mater. 2004;3:249–253. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- 14.Park J, Kalinin YV, Kadam S, Randall CL, Gracias DH. Artif. Organs. 2013;37:1059–1067. doi: 10.1111/aor.12131. [DOI] [PubMed] [Google Scholar]
- 15.Owen SC, Shoichet MS. J. Biomed. Mater. Res., Part A. 2010;94:1321–1331. doi: 10.1002/jbm.a.32834. [DOI] [PubMed] [Google Scholar]
- 16.Burdick JA, Khademhosseini A, Langer R. Langmuir. 2004;20:5153–5156. doi: 10.1021/la049298n. [DOI] [PubMed] [Google Scholar]
- 17.Therriault D, White SR, Lewis JA. Nat. Mater. 2003;2:265–271. doi: 10.1038/nmat863. [DOI] [PubMed] [Google Scholar]
- 18.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DH, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Nat. Mater. 2012;11:768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chien MP, Rush AM, Thompson MP, Gianneschi NC. Angew. Chem., Int. Ed. 2010;49:5076–5080. doi: 10.1002/anie.201000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee MH, Hribar KC, Brugarolas T, Kamat NP, Burdick JA, Lee D. Adv. Funct. Mater. 2012;22:131–138. [Google Scholar]
- 21.Randolph LM, Chien MP, Gianneschi NC. Chem. Sci. 2012;3:1363–1380. doi: 10.1039/C2SC00857B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson MP, Chien MP, Ku TH, Rush AM, Gianneschi NC. Nano Lett. 2010;10:2690–2693. doi: 10.1021/nl101640k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohamed F, van der Walle CF. J. Pharm. Sci. 2008;97:71–87. doi: 10.1002/jps.21082. [DOI] [PubMed] [Google Scholar]
- 24.Shah RK, Shum HC, Rowat AC, Lee D, Agresti JJ, Utada AS, Chu L-Y, Kim J-W, Fernandez-Nieves A, Martinez CJ, Weitz DA. Mater. Today. 2008;11:18–27. [Google Scholar]
- 25.Utada AS, Lorenceau E, Link DR, Kaplan PD, Stone HA, Weitz DA. Science. 2005;308:537–541. doi: 10.1126/science.1109164. [DOI] [PubMed] [Google Scholar]
- 26.Xu S, Nie Z, Seo M, Lewis P, Kumacheva E, Stone HA, Garstecki P, Weibel DB, Gitlin I, Whitesides GM. Angew. Chem. 2005;117:734–738. doi: 10.1002/anie.200462226. [DOI] [PubMed] [Google Scholar]
- 27.Bai S, Debnath S, Gibson K, Schlicht B, Bayne L, Zagnoni M, Ulijn RV. Small. 2014;10:285–293. doi: 10.1002/smll.201301333. [DOI] [PubMed] [Google Scholar]
- 28.Shah RK, Kim JW, Weitz DA. Langmuir. 2010;26:1561–1565. doi: 10.1021/la9041327. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Byrne JD, Napier ME, DeSimone JM. Adv. Drug Delivery Rev. 2012;64:1021–1030. doi: 10.1016/j.addr.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian L, Gandra N, Singamaneni S. ACS Nano. 2013;7:4252–4260. doi: 10.1021/nn400728t. [DOI] [PubMed] [Google Scholar]
- 31.Timko BP, Arruebo M, Shankarappa SA, McAlvin JB, Okonkwo OS, Mizrahi B, Stefanescu CF, Gomez L, Zhu J, Zhu A, Santamaria J, Langer R, Kohane DS. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1349–1354. doi: 10.1073/pnas.1322651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mannoor MS, Jiang Z, James T, Kong YL, Malatesta KA, Soboyejo WO, Verma N, Gracias DH, McAlpine MC. Nano Lett. 2013;13:2634–2639. doi: 10.1021/nl4007744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis JA. Adv. Funct. Mater. 2006;16:2193–2204. [Google Scholar]
- 34.Barry RA, Shepherd RF, Hanson JN, Nuzzo RG, Wiltzius P, Lewis JA. Adv. Mater. 2009;21:2407–2410. [Google Scholar]
- 35.Wu W, DeConinck A, Lewis JA. Adv. Mater. 2011;23:H178–H183. doi: 10.1002/adma.201004625. [DOI] [PubMed] [Google Scholar]
- 36.Hanson Shepherd JN, Parker ST, Shepherd RF, Gillette MU, Lewis JA, Nuzzo RG. Adv. Funct. Mater. 2011;21:47–54. doi: 10.1002/adfm.201001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong YL, Tamargo IA, Kim H, Johnson BN, Gupta MK, Koh TW, Chin HA, Steingart DA, Rand BP, McAlpine MC. Nano Lett. 2014;14:7017–7023. doi: 10.1021/nl5033292. [DOI] [PubMed] [Google Scholar]
- 38.Ladd C, So JH, Muth J, Dickey MD. Adv. Mater. 2013;25:5081–5085. doi: 10.1002/adma.201301400. [DOI] [PubMed] [Google Scholar]
- 39.Pataky K, Braschler T, Negro A, Renaud P, Lutolf MP, Brugger J. Adv. Mater. 2012;24:391–396. doi: 10.1002/adma.201102800. [DOI] [PubMed] [Google Scholar]
- 40.Willets KA, Van Duyne RP. Annu. Rev. Phys. Chem. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- 41.Um E, Rogers ME, Stone HA. Lab Chip. 2013;13:4674–4680. doi: 10.1039/c3lc50957e. [DOI] [PubMed] [Google Scholar]
- 42.Majd S, Mayer M. Angew. Chem., Int. Ed. 2005;44:6697–6700. doi: 10.1002/anie.200502189. [DOI] [PubMed] [Google Scholar]
- 43.Frederix PWJM, Scott GG, Abul-Haija YM, Kalafatovic D, Pappas CG, Javid N, Hunt NT, Ulijn RV, Tuttle T. Nat. Chem. 2015;7:30–37. doi: 10.1038/nchem.2122. [DOI] [PubMed] [Google Scholar]
- 44.Huang X, Brazel CS. J. Controlled Release. 2001;73:121–136. doi: 10.1016/s0168-3659(01)00248-6. [DOI] [PubMed] [Google Scholar]
- 45.Tian L, Chen E, Gandra N, Abbas A, Singamaneni S. Langmuir. 2012;28:17435–17442. doi: 10.1021/la3034534. [DOI] [PubMed] [Google Scholar]
- 46.Sherry LJ, Jin R, Mirkin CA, Schatz GC, Van Duyne RP. Nano Lett. 2006;6:2060–2065. doi: 10.1021/nl061286u. [DOI] [PubMed] [Google Scholar]
- 47.Maity S, Wu WC, Xu C, Tracy JB, Gundogdu K, Bochinski JR, Clarke LI. Nanoscale. 2014;6:15236–15247. doi: 10.1039/c4nr05179c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jain PK, Lee KS, El-Sayed IH, El-Sayed MA. J. Phys. Chem. B. 2006;110:7238–7248. doi: 10.1021/jp057170o. [DOI] [PubMed] [Google Scholar]
- 49.Incropera FP. Fundamentals of Heat and Mass Transfer. New York: John Wiley & Sons; 2011. [Google Scholar]
- 50.Blumm J, Lindemann A. High Temp.—High Pressures. 2003/2007;35/36:627–632. [Google Scholar]
- 51.Henzie J, Lee MH, Odom TW. Nat. Nanotechnol. 2007;2:549–554. doi: 10.1038/nnano.2007.252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.