Abstract
In Arabidopsis, various cell type-specific whole-genome expression analyses have been conducted. However, the vast majority of these were performed with cellular RNA from root tissues or other easily accessible cell types [1]. Nuclear RNA was neglected for a long time as not being representative for transcriptomic studies. In recent years, however, there have been reports describing the validity of nuclear RNA for these types of studies [2], [3]. Here we describe the generation, quality assessment and analysis of nuclear transcriptomic data from Arabidopsis embryos published by Slane et al. (2014) [4]. Comparison of nuclear with cellular gene expression demonstrated the usefulness of nuclear transcriptomics.
Keywords: Arabidopsis thaliana, Gene expression, Microarray, Nuclear and cellular transcriptome, Pro-embryo and suspensor transcriptome
| Specifications | |
|---|---|
| Organism/tissue | Arabidopsis thaliana/embryo |
| Sex | N/A |
| Sequencer or array type | Affymetrix GeneChip® Arabidopsis ATH1 Genome Array |
| Data format | Raw data: CEL files |
| Experimental factors | Cell-type specific transcriptomes of pre-globular pro-embryo and suspensor |
| Experimental features | Transcriptome analysis to study comparability of nuclear and cellular expression |
| Consent | N/A |
Direct link to deposited data
Experimental design, materials and methods
Manual embryo dissection and isolation of GFP-labeled nuclei
For manual embryo dissection, Arabidopsis ovules were dissected out of siliques and intact embryos at early globular stage (16 cells and 32 cells) were squeezed out of the ovules on a microscopic slide by gently applying pressure to a cover slip. Afterwards, released embryos were manually collected by pipetting using 10 μl pipette tips rinsed with 1% BSA (bovine serum albumin). Embryos were thoroughly washed twice in water to remove adhering particles from the endosperm and maternal seed tissue. During the course of embryo isolation and prior to RNA extraction, embryos were stored in RNAlater (Qiagen). For isolation of GFP-labeled nuclei prior to fluorescence-activated nuclear sorting (FANS), approximately 100 young ovules containing embryos at earliest embryonic stages (1 cell to 16 cells) were collected in 50 μl RNAlater. Afterwards, 1.5 μl 4% paraformaldehyde (PFA) solved in phosphate buffered saline (PBS) was added to a final concentration of approximately 0.12% and the ovules were incubated for 5 min at room temperature. The fixed tissue was thoroughly homogenized with a plastic pestle in a 1.5 ml tube and nuclear extraction was performed with slight modifications according to the crude preparation protocol using the CelLytic™ PN kit (Sigma-Aldrich). Since it was possible to completely homogenize the tissue samples by grinding, no filtering step was applied prior to the following centrifugation steps. The cell membrane lysis was aided by incubation with Triton X-100 0.3% for 10 min. Importantly, tissue pellets were dissolved as completely as possible before every centrifugation step by pipetting and thorough vortexing. The GFP-positive nuclei were then separated from the total pool of isolated nuclei by fluorescence-activated nuclear sorting (FANS) using a MoFlo Legacy (Beckman Coulter) FACS and collected in 150 μl freshly prepared Proteinase K buffer (10 mM Tris–HCl (pH 7.9), 50 mM EDTA (pH 7.9), 0.2 M NaCl, 0.5% SDS, 0.5 mg/ml RNase inhibitor (Fermentas), 600 μg/ml Proteinase K) [5].
RNA extraction, quality control and RNA amplification
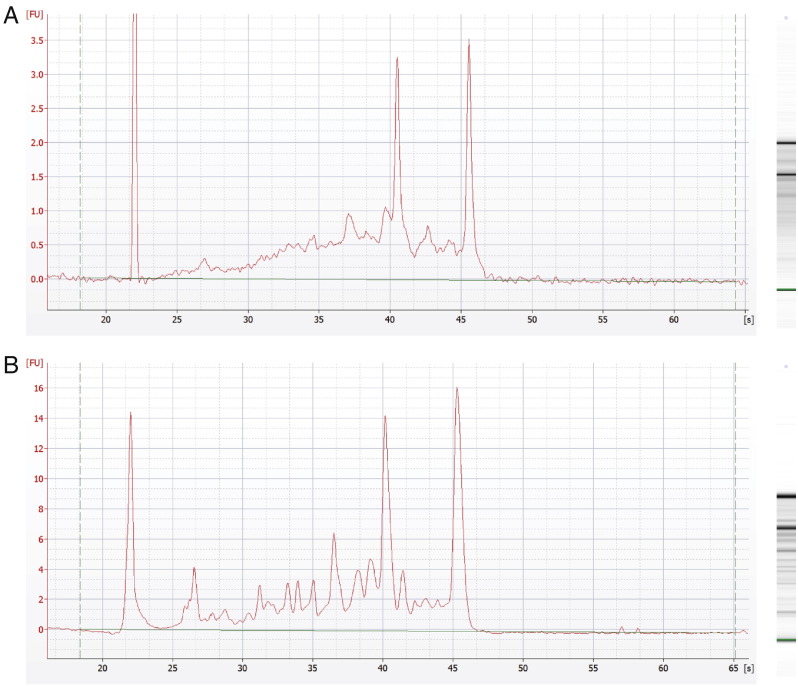
RNA from manually collected embryos was extracted with the RNeasy Micro Kit (QIAGEN). The Proteinase K buffer containing isolated nuclei was incubated at 55 °C for 10–15 min with intense shaking. All following steps were performed at 4 °C and centrifugation steps were carried out at 14,000 g. RNase-free water to 600 μl and an equal volume of phenol (pH 4.2) was added, thoroughly mixed and spun for 10 min. The aqueous phase was transferred to a new tube and an equal volume of phenol:chloroform (1:1) was added. After thorough mixing and centrifugation for 10 min the aqueous phase was transferred to a new tube and an equal volume of pre-cooled isopropanol and 20 μg glycogen was added. The sample was then precipitated for at least 30 min at − 20 °C and spun for 30–45 min. The pellet was subsequently washed with 600 μl pre-cooled 70% ethanol and another centrifugation step followed for 10 min. The pellet was air-dried and dissolved in RNase-free water. DNase I treatment was mainly performed on-column according to the kit protocol (QIAGEN). Prior to quality control and amplification, the RNA was concentrated with a vacuum cooling centrifuge. Further, the quality of the extracted nuclear and cellular RNA was assessed on a RNA pico chip with a 2100 Bioanalyzer (Agilent technologies) (Fig. 1). To increase the similarity between nuclear and cellular transcriptomes, we ensured that only nuclear poly-adenylated mRNAs ready for export to the cytoplasm were profiled by using a polyT primer-based linear RNA amplification kit (RiboAmp HS PLUS RNA Amplification Kit, Arcturus). Starting from an average of 2 ng total nuclear RNA per biological replicate, after 1.5 rounds of amplification (total RNA → cDNA → amplified RNA → cDNA) the amplified cDNA was further amplified and concomitantly labeled with biotin by another round of reverse transcription (BioArray Single-round RNA amplification and biotin labeling system, ENZO Life Sciences). Also for the cellular RNA from dissected embryos, 1.5 rounds of RNA amplification were employed in a similar fashion.
Fig. 1.
Quality analysis of extracted RNA. Electropherogram and gel picture for nuclear (A) and cellular (B) RNA.
Microarray expression data analysis and comparison
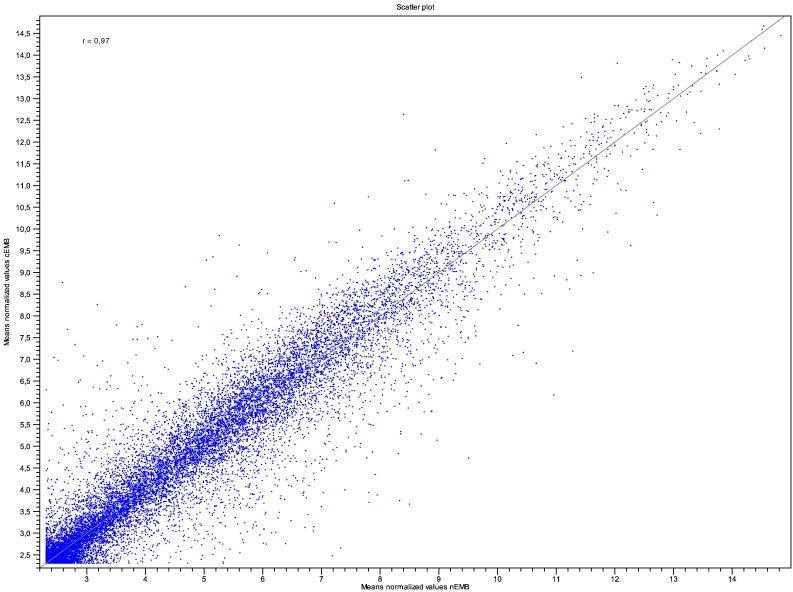
For expression analysis, GeneChip® Arabidopsis ATH1 Genome Arrays (Affymetrix) were used. RNA fragmentation and hybridization to the microarray, washing and staining steps as well as array scanning were performed according to the Affymetrix standard protocol. Microarray data analysis and normalization of raw values was carried out with different packages implemented in ‘R’ (v2.14.2; http://www.r-project.org/). Log2-based transformation of values from CEL files was achieved using ‘gcRMA’ (v2.26.0) [6]. Means of gcRMA transformed values for three microarray replicates for embryonic nuclear (nEMB) and cellular (cEMB) RNA were compared using CLC Main Workbench version 7.6. As revealed by the correlation scatter plot, there is a high correlation between the expression data obtained from nuclear and cellular embryonic transcriptomes, indicating that nuclear poly-adenylated transcripts can be used as a reliable proxy to determine cellular mRNA abundance (Fig. 2). Subsequently, we used this technique to obtain cell type-specific transcriptome data from pro-embryo and suspensor cells, two inaccessible yet distinct tissue types [4].
Fig. 2.
Correlation analysis of nuclear and cellular expression values. Correlation scatter plot comparing means of log2-transformed expression estimates based on three biological replicates each. Additionally, R value is indicated.
Discussion
In Arabidopsis, most cell type-specific whole genome expression studies are performed with cellular mRNA [1]. However, for difficult-to-access tissue utilizing nuclear RNA can be advantageous [2], [3]. We describe here in detail the generation of gene-expression data from nuclear RNA as well as cellular total RNA of Arabidopsis early embryos. As was shown in the aforementioned publication by Slane et al., there is high correlation between the poly-adenylated RNA populations of nuclei and whole cells. This demonstrates that nuclear RNA can be reliably used for transcriptome analyses of specific cell-types, especially in tissues, which are not easily accessible otherwise. Furthermore, this technique could also be used for DNA isolation and should in principle enable the analysis of tissue-specific chromatin modifications.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
This work was funded by the SIREN network [European Commission under FP7-PEOPLE-2007-1-1-ITN to G.J.], the DFG [Deutsche Forschungsgemeinschaft, BA3356/2-1 and SFB1101 to M.B.] and the Max Planck Society.
References
- 1.Palovaara J., Saiga S., Weijers D. Transcriptomics approaches in the early Arabidopsis embryo. Trends Plant Sci. 2013;18:514–521. doi: 10.1016/j.tplants.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 2.Barthelson R.A., Lambert G.M., Vanier C., Lynch R.M., Galbraith D.W. Comparison of the contributions of the nuclear and cytoplasmic compartments to global gene expression in human cells. BMC Genomics. 2007;8:340. doi: 10.1186/1471-2164-8-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang C., Barthelson R.A., Lambert G.M., Galbraith D.W. Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol. 2008;147:30–40. doi: 10.1104/pp.107.115246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slane D., Kong J., Berendzen K.W., Kilian J., Henschen A., Kolb M., Schmid M., Harter K., Mayer U., De Smet I., Bayer M., Jürgens G. Cell type-specific transcriptome analysis in the early Arabidopsis thaliana embryo. Development. 2014;141:4831–4840. doi: 10.1242/dev.116459. [DOI] [PubMed] [Google Scholar]
- 5.Khodosevich K., Inta D., Seeburg P.H., Monyer H. Gene expression analysis of in vivo fluorescent cells. PLoS ONE. 2007;2:e1151. doi: 10.1371/journal.pone.0001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z., Irizarry R.A., Robert G., Martinez-Murillo F., Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909–917. [Google Scholar]