Abstract
Regulatory T cells (Treg) play a central role in the suppression of inflammatory and allergic responses. Colonization of certain gut commensal microbes such as Clostridia class IV and XIVa in the gut can induce development of colonic Treg cells contributing to the maintenance of gut immune homeostasis. Clostridia-derived butyrate promotes the differentiation of naïve T cells into Treg cells through upregulation of Foxp3, the master transcription factor of Treg cells. Chromatin immunoprecipitation-sequencing (ChIP-seq) analysis revealed that treatment of naïve T cells with butyrate induces Treg-polarizing conditions by enhanced histone H3 acetylation in the promoter and conserved non-coding sequence regions of the Foxp3 locus. In general, global normalization was utilized for ChIP-seq analysis to compare the data obtained from two or more samples. However, global normalization is not appropriate for the evaluation of ChIP-seq data when treatment can affect the total amount of target protein. Here, we introduce a unique normalization method for ChIP-seq analysis in cells treated with butyrate, a pan-HDAC inhibitor that is likely to affect total acetylation levels of histone H3.
Keywords: ChIP-seq, Microarray, Butyrate, HDAC, Normalization, Regulatory T cells
| Specifications | |
|---|---|
| Organism/cell line/tissue | Mus musculus/splenic naive CD4+ T cells |
| Sex | Male |
| Sequencer or array type | Hi-seq1000 (Illumina) GeneChip® Mouse Gene 1.0 ST Array qRT-PCR |
| Data format | Raw and processed |
| Experimental factors | Cells |
| Experimental features | Cells in the presence or absence of butyrate under Treg-inducing condition |
| Consent | n/a |
| Sample source location | n/a |
Direct link to deposited data
The microarray and ChIP-seq analysis data were deposited in the Gene Expression Omnibus (GEO): http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49655.
Materials and methods
Cell culture
The current Materials and methods section was basically described in our previous paper [1]. CD4+ T cells were enriched from the spleen and lymph nodes of C57BL/6 mice by a negative selection method with the IMag Cell Separation System (BD Bioscience, San Jose, CA) using a mixture of biotinylated mouse monoclonal antibodies (mAbs) against B220 (RA3-6B2), CD8 (53–6.7), CD11c (N418), Gr-1 (RB6-8C5), Ter-119 (TER-119), and Streptavidin Particle Plus-DM (all from BD Biosciences). The enriched CD4+ fraction was subjected to cell sorting with FACSAriaII to isolate CD3+CD4+CD25−CD44loCD62Lhi naive T cells using anti-CD3-FITC, (145-2C11), anti-CD4-PE (GK1.5), anti-CD25-APC (PC61), anti-CD44-PECy7 (IM7), anti-CD62L-PB (MEL-14) mAbs. For Treg polarization, naive CD4+ T cells (5 × 105 cells/ml, 48-well plate) were stimulated with immobilized anti-CD3 mAb (145-2C11, without Sodium Azide, 10 μg/ml) and soluble anti-CD28 mAb (37.51, without Sodium Azide, 1 μg/ml) supplemented with 0.2 ng/ml TGF-β1 and 10 ng/ml IL-2 (both from R&D Systems) in the presence or absence of 0.1 mM sodium butyrate (Sigma-Aldrich), a well-known pan-HDAC inhibitor [2]. Cells subjected to ChIP-seq analysis were cultured for 1 day, and cells subjected to microarray analysis were cultured for 2 days.
RNA isolation & quantitative PCR
Total RNA was extracted by TRIzol reagent (Life Technologies, Carlsbad, CA) following a standard protocol. RNA quality was analyzed using a Bioanalyzer 2100 and the RNA6000 Nano LabChip kit (Agilent Technologies, Inc., Santa Clara, CA). RNA samples that had RNA integrity number (RIN) values above 9.5 were considered acceptable.
Microarray analysis
Gene expression was analyzed using a GeneChip® Mouse Gene 1.0 ST Array spotted with approximately 26,166 probe sets (Affymetrix, Santa Clara, CA). Samples for array hybridization were prepared as described in the Affymetrix GeneChip® Expression Technical Manual. The scanned arrays were analyzed using the GeneChip® Analysis Suite software (Affymetrix). The obtained hybridization intensity data and qualities were checked using the GeneSpring® analysis software (Silicon Genetics, Redwood City, CA).
Fixation and chromatin shearing
For chromatin immunoprecipitation-sequencing analysis (ChIP), 5 × 105 cells were resuspended with 500 μL of phosphate-buffered saline (PBS) at room temperature (RT) and then fixed with 13.5 μL of 37% formaldehyde (RT, 10 min) and the reaction was stopped by the addition of 1 M glycine to a final concentration of 125 mM (RT, 5 min). Cells were washed twice with ice-cold PBS, frozen in liquid nitrogen, and stored at − 80 °C until analysis. The frozen cell pellet was re-suspended with 20 μL SDS lysis buffer (Life Technologies), on ice for 10 min to obtain the chromatin solution. The chromatin solution was diluted up to 130 μL with dilution buffer (Life Technologies), transferred to AFA microtubes, and sonicated using a focused-ultrasonicator (Covaris S2, Covaris, MA) to obtain the fragmented chromatin DNA. The acoustic parameters were optimized (Duty cycle: 10%, Intensity: 5, Cycle/Burst: 200, 15 min, < 8 °C) to reduce the DNA length to approximately 100–300 bp. The power output was much higher than that recommended by the kit manufacturer but the appropriate conditions for fragmentation seems to be dependent on the fixation time and cell type. The size of the DNA fragment was monitored using an Agilent 2100 Bioanalyzer and the High Sensitivity DNA kit.
ChIP-seq analysis
ChIP assays were performed using the MAGnify ChIP system (Life Technologies, NY) according to the manufacturer's protocol with a few modifications. The sheared chromatin was divided into two tubes, adjusted to 150 μL with dilution buffer, and immunoprecipitated (overnight, 4 °C) with magnetic protein A/G beads with 3 μg of immobilized anti-acetyl histone H3 polyclonal antibody (Millipore) or rabbit IgG on a rotating wheel. The beads were washed with IP1 buffer (3 times) and IP2 buffer (twice). Immune complexes were eluted (55 °C, 30 min) and reverse crosslinking was carried out (65 °C, 1 h) in the presence of Proteinase K. Quantitative real-time PCR analysis was performed following magnetic bead-based DNA purification. For ChIP-seq library construction, immunoprecipitated DNA fragments were ligated with adaptors and amplified by PCR using NEBNext Ultra DNA Library Prep kit from Illumina. Because the amount of precipitated DNA was too low (approximately 1 ng per IP) compared with the manufacturer recommendations, PCR cycles were increased to 18 so that an adequate amount of DNA was obtained for cluster generation. The number of PCR cycles is based on the protocol for ChIP-seq analysis provided by Illumina (ChIP-seq Sample Prep Kit for Illumina). Probably due to the low amount of input DNA, excess short DNA fragments, approximately 80 bp arising from primer dimerization and 120 bp from adapter–adapter ligation, can be detected in most cases. DNA fragments of an undesirable length were removed using a 2% E-gel system (Life Technologies). Amplified libraries were quantified using the KAPA Library Quantification Kit following measurement of the average DNA length using the Agilent 2100 Bioanalyzer. Library concentration was adjusted to 2 nM and 10 μL of the libraries were subjected to cluster generation and sequencing analysis with the Hi-Seq 1000 (50 bp single-end run, Illumina). Sequenced reads were mapped to the mouse genome (ver. mm9) with BOWTIE.
ChIP-qPCR analysis
The DNA samples were amplified with the Eco Real Time PCR System (Illumina) using SYBR premix Ex Taq (TAKARA BIO) and the primer sets specific for mouse genes. The sequences of the primer sets used for ChIP-seq normalization are listed in Table 1.
Table 1.
Primer sequences for ChIP-qPCR of Rpl13a gene loci.
| 60S ribosomal protein L13a (Rpl13a) gene (52380933–52384115) | |
| Promoter | |
| − 700 – − 833 bp | |
| C57BL/6 J chromosome 52384815–52384948 | |
| Forward primer | ACGCCCCATGGGAATTTGAT |
| Reverse primer | AAAACCTCCCTAACCAGCCG |
| Product length | 134 |
| − 722 – − 796 bp | |
| C57BL/6 J chromosome 7, 52384837–52384911 | |
| Forward primer | TTCCCCAAAACCATAGCCCC |
| Reverse primer | CCTGAGTCGAGTCCTCCTGT |
| Product length | 75 |
| + 219 – + 310 bp | |
| C57BL/6 J chromosome 7, 52383806–52383878 | |
| Forward primer | AAATGCAGCGGGTTTTCTGG |
| Reverse primer | CCCCAACGCCTGAGATTACA |
| Product length | 92 |
| + 293 – + 414 bp | |
| C57BL/6J chromosome 7, 52383702–52383823 | |
| Forward primer | TAATCTCAGGCGTTGGGGTG |
| Reverse primer | TACAGGAATCCGGTGCCCTA |
| Product length | 122 |
Global normalization and data analysis
Microarray signals were processed using a standard RMA algorithm. Observed signals were normalized using a quantile normalization method and genes that had no significant signals were ignored to reduce noise.
ChIP peaks for each population were called with MACS with p value threshold of less than 10− 5. The numbers recorded for reads in a section were that of total mapped reads and the length of each section. The genes that are differentially histone H3-acetylated between conditions were identified using the normalized reads, which mapped from 4-kb upstream to 4-kb downstream of the transcription start site (TSS). TSS regions were defined based on annotation in the Entrez database.
Integrative analysis of ChIP-seq data and microarray data
Transcription factors were selected using annotation obtained from the database Gene Ontology (http://geneontology.org/). We assumed that transcription factors possess both the “nucleus (GO:0005634)” and “regulation of transcription, DNA-templated (GO:0006355)” GO terms.
Among 1350 genes, 663 transcription factor genes were identified as butyrate sensitive genes based on differences in histone acetylation levels with or without butyrate treatment. The distribution of the log ratio of promoter acetylation was fitted with two Gaussian curves using the EM algorithm. We determined the threshold where the false discovery rate (FDR) in the upper group was 0.05, meaning less than 1 of 20 or > 5% of the identified genes were likely to have been incorrectly included in the group.
Expression changes in transcription factors after butyrate treatment were calculated based on the microarray analysis. Log ratios of expression changes between the two groups were measured and compared by the Student's t-test.
Normalization of ChIP-seq data by Rpl13a in cells treated with butyrate
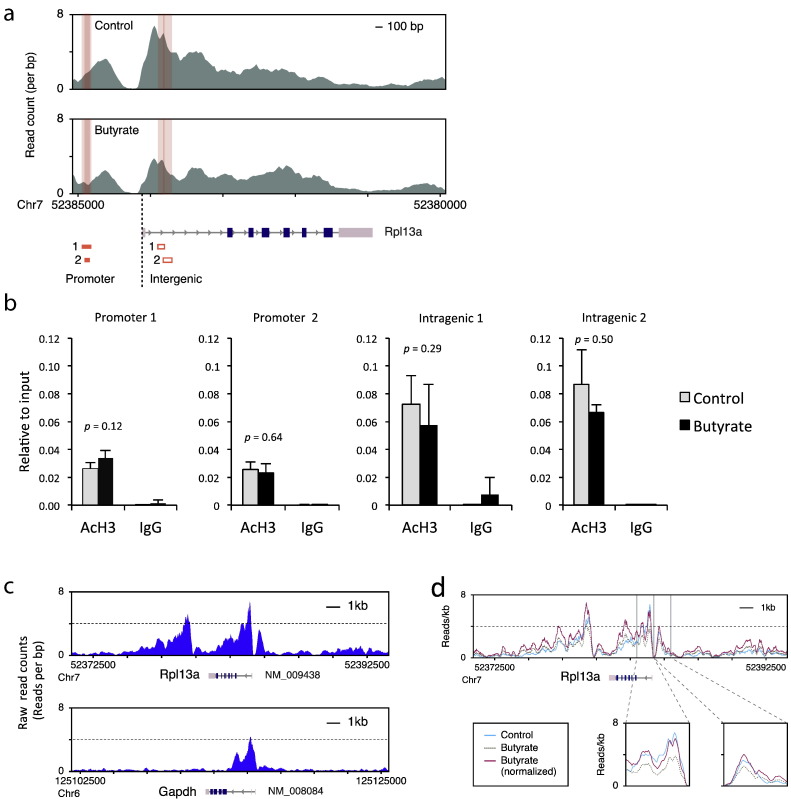
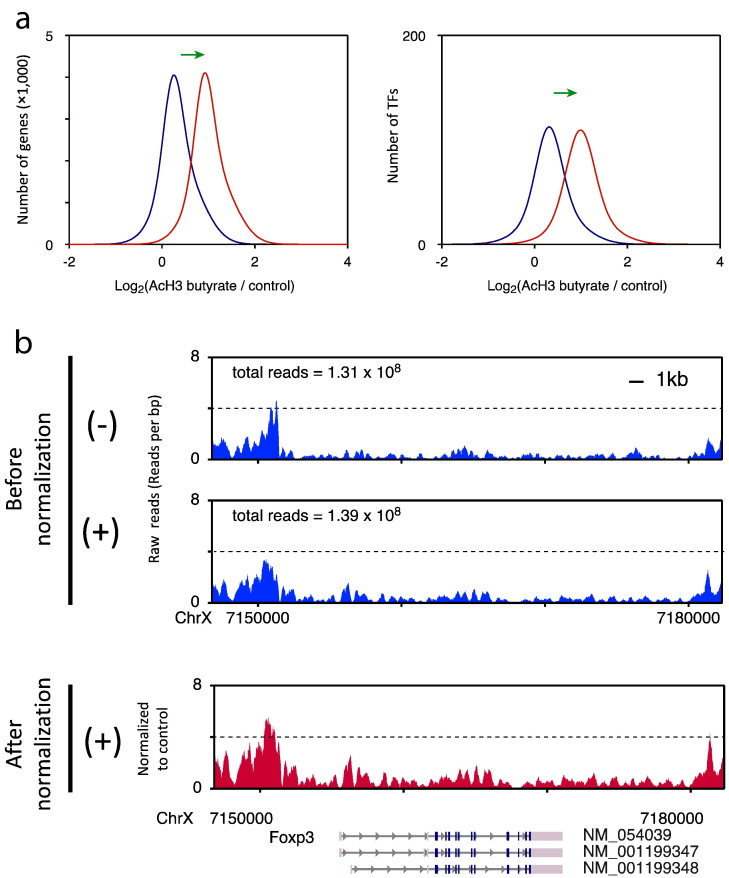
Because butyrate treatment globally enhanced histone acetylation (Supplementary Fig. 20 in [1]), total mapped reads may be increased in butyrate-treated cells compared to control cells, leading to an underestimation of histone acetylation changes for all gene loci in cells treated with butyrate [3]. In fact, read counts on Rpl13a promoters and genes in ChIP-seq analysis did not correlate with the proportion of enrichment in ChIP-qPCR analysis (Fig. 1a, b). The lack of correlation between ChIP-seq data and ChIP-qPCR data for the same samples is probably due to the global normalization of ChIP-seq data. Butyrate increases the acetylation of histone H3 [1], [2], [3]; however, the global normalization method assumes that there is no significant difference between the total histone H3 levels of samples treated with and without butyrate. Therefore, the normalized values within each sample need to be adjusted based on the read counts at the four genomic regions around the TSS of the housekeeping gene Rpl13a. The Rpl13a gene is suitable for normalization of other genes, because the expression level of this gene is more stable than Gapdh and ActB under various conditions [4]. In addition, the histone acetylation levels at the Rpl13a promoter and intragenic regions are most likely saturated and are much higher than that of Gapdh in cultured T cells (Fig. 1c). Therefore, we expected that the histone acetylation status of Rpl13a probably remained unchanged between butyrate-treated and untreated cells. Indeed, we confirmed this expectation by ChIP-qPCR analysis (Fig. 1d). These primers were designed to target the peaks before and after the TSS, where the peak heights were almost the highest in the promoter and the intragenic region of Rpl13a. The normalized reads were calculated within a fixed base length for the upstream and downstream regions, and a relative length for genic regions, as follows: the number of reads in promoter regions and intragenic regions were counted as shown in Table 2; the number of reads in the Rpl13a promoter and intragenic regions of control cells were divided by those in butyrate-treated cells; and the log2 ratios were compared to those obtained from the ChIP-qPCR data. Based on the difference between the 4 regions of Rpll3a in the ChIP-seq and ChIP-qPCR data, we calculated that the number of reads in butyrate-treated cells should be multiplied by 1.686. By using this normalization method, underestimation of the histone acetylation changes on the promoter regions of Rpl13a and other all genes, including Foxp3, in butyrate treated cells can be rectified (Fig. 1d and Fig. 2a, b). It might be possible to use other genes for normalization if their expression levels and histone acetylation levels were almost saturated in target cells. Notably, we should be careful when normalizing histone acetylation levels not only in cells treated with pan-HDAC inhibitors but also in cells under starvation, because starvation may affect expression levels of housekeeping genes such as ActB and Gapdh [5].
Fig. 1.
ChIP-seq (a) and ChIP-qPCR (b) analysis of promoter and intragenic regions of Rpl13a was performed in CD4+ T cells cultured in the presence or absence of butyrate for 1 day, and immunoprecipitated using anti-acetyl histone H3 antibody (AcH3) or rabbit IgG as a negative control. Shading in Fig. 1a shows the ChIP-seq peaks. Orange boxes in Fig. 1a indicate the regions corresponding to the qPCR products in Fig. 1b. The error bars indicate standard deviation (n = 3). Comparison of peak height in Rpl13a and Gapdh genes in control cells (c). Normalized or raw peak height in Rpl13a genes in cells with or without butyrate (d).
Table 2.
Comparison of the number of reads in ChIP-seq analysis with ChIP-qPCR data of Rpl13a gene loci.
| ChIP-seq analysis |
ChIP-qPCR analysis |
||||||
|---|---|---|---|---|---|---|---|
| Name | Location | Control (reads) | Butyrate (reads) | Ratio (ChIP-seq) | Log2 (ChIP-seq, ratio) | ChIP-qPCR | Log2 (ChIP-qPCR) |
| Promoter1 | chr7:52384815:52384948 | 303 | 154 | 1.97 | 0.98 | 1.274 | 0.349 |
| Promoter2 | chr7:52384837:52384911 | 213 | 123 | 1.73 | 0.79 | 0.886 | − 0.175 |
| Intragenic1 | chr7:52383806:52383897 | 436 | 215 | 2.03 | 1.02 | 0.794 | − 0.333 |
| Intragenic2 | chr7:52383702:52383823 | 585 | 348 | 1.68 | 0.75 | 0.777 | − 0.364 |
| Correction value | 1.686 | ||||||
Fig. 2.
ChIP-seq analysis of the promoter regions of all genes and transcription factors including Foxp3 with (red line) or without (blue line) normalization based on histone acetylation levels in Rpl13a loci. The distribution of acetylated H3 peak signals (a), green arrow indicates the application of Rpl13a-based normalization. Acetylated histone H3 levels in the Foxp3 gene locus with (−) or without (+) butyrate treatment (b).
References
- 1.Furusawa Y., Obata Y., Fukuda S. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 2.Candido E.P., Reeves R., Davie J.R. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978;14:105–113. doi: 10.1016/0092-8674(78)90305-7. [DOI] [PubMed] [Google Scholar]
- 3.Rada-Iglesias A., Enroth S., Ameur A. Butyrate mediates decrease of histone acetylation centered on transcription start sites and down-regulation of associated genes. Genome Res. 2007;17:708–719. doi: 10.1101/gr.5540007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Jonge H.J.M., Fehrmann R.S.N., de Bont E.S.J.M. Evidence based selection of housekeeping genes. PLoS ONE. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimittgen T.D., Zakrajsek B.A. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods. 2000;20:1–13. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]