SUMMARY
Adeno-associated viral vectors (AAV) have emerged as a gene delivery platform with demonstrated safety and efficacy in a handful of clinical trials for monogenic disorders. However, limitations of the current generation vectors often prevent broader application of AAV gene therapy. Efforts to engineer AAV have been hampered by a limited understanding of the structure-function relationship of the complex multimeric icosahedral architecture of the particle. To develop additional reagents pertinent to further our insight into AAV, we inferred evolutionary intermediates of the viral capsid using ancestral sequence reconstruction. In silico derived sequences were synthesized de novo and characterized for biological properties relevant to clinical applications. This effort led to the generation of 9 functional putative ancestral AAVs and the identification of Anc80, the predicted ancestor of the widely studied AAV serotypes 1, 2, 8 and 9 as a highly potent in vivo gene therapy vector for targeting liver, muscle, and retina.
Keywords: Gene Transfer, AAV, adeno-associated virus, vector, gene therapy, ancestral reconstruction, virus, synthetic biology, neutralizing antibody, pre-existing immunity, capsid T-cell, vaccine
INTRODUCTION
Extensive preclinical studies have established a favorable safety profile for adeno-associated viral vector (AAV). In addition, AAV has enabled demonstration of in vivo gene therapy efficacy in animal models of disease with etiologies ranging from inherited, infectious to common complex (Hastie and Samulski, 2015; Schnepp and Johnson, 2014). Furthermore, recent early stage AAV clinical trials have led to the first demonstrations of clinical benefit in 2 forms of inherited blindness with AAV2 (Bainbridge et al., 2008; Jacobson et al., 2012; MacLaren et al., 2014; Maguire et al., 2008) and hemophilia B with AAV8 (Nathwani et al., 2011). One treatment based on AAV1 has been awarded a drug license by European regulators (Bryant et al., 2013). Based on these data, AAV has been proposed as a platform technology for therapeutic in vivo gene delivery.
AAV is a 25 nm non-enveloped icosahedral capsid virus carrying a 4.7 kb ssDNA genome flanked by inverted terminal repeats (ITRs). AAV classifies as a Dependoparvovirus genus within the Parvoviridae family. Its genome comprises genes encoding for replication (Rep), structural capsid (Cap) and assembly (AAP) proteins. AAV is a helper-dependent virus, requiring the heterologous cofactors to complete a replicative cycle that an adenovirus or herpesvirus can provide in the context of a co-infection. Replication deficient AAV can be generated by eliminating all viral coding sequence in cis, and providing those in trans during vector production. Particles generated in this manner can encode any type of transgene cassette that does not exceed the size of the genome of the wild type virus (approximately 4.7 kb) and enable gene transfer in vitro and in vivo to multiple cell and tissue targets (Knipe and Howley, 2013).
The AAV particle is composed of three C-terminally overlapping Cap proteins named VP1, 2, and 3. VP3, the smallest member of these structural proteins is necessary and sufficient for full capsid assembly through multimerization. VP1, required for particle infectivity, and VP2, reported to be redundant structurally or functionally, are embedded in the wild type T=1 viral architecture at a VP1:2:3 ratio of approximately 1:1:10. A total of 60 VP monomers assemble into a icosahedral capsomer along 2, 3, and 5-fold axes of symmetry (Knipe and Howley, 2013). Every monomer within the 60-mer structure interfaces with 7 neighboring capsid monomers. High resolution crystallography studies identify a conserved core structure composed of an eight-stranded β-barrel motif and the αA helix as well as nine surface exposed domains (VR-I to -IX) which can vary between the known primate AAV serotypes (DiMattia et al., 2012; Xie et al., 2002).
The structural diversity of natural AAVs is thereof largely contained within the nine surface regions of the capsid, which functionally results in divergent receptor binding properties, post-entry trafficking, host response, and gene transfer efficiency to various cell and tissue targets. Structural determinant for many of these properties have largely remained elusive with a few notable exceptions: receptor binding motifs for certain serotypes (Bell et al., 2012; Kern et al., 2003; Wu et al., 2000), phospholipase activity and nuclear localization activity on VP1,2 unique domains (Girod et al., 2002; Grieger et al., 2006), a limited number of docking sites for monoclonal antibodies (DiMattia et al., 2012), a motif enhancing T-cell immunogenicity (Vandenberghe et al., 2006), and a select number MHCI restricted T-cell epitopes (Mingozzi et al., 2007) have previously been mapped..
In an era of active clinical translation in AAV gene therapy, there is a desire to better understand the structure-function relationship of AAV within the constraints of the particle architecture in order to further model and modulate the pharmacology of this new class of drugs to improve transduction efficiency and specificity, alter tropism, and reduce immunogenicity. To enable mechanistic studies, map structural determinants of these phenotypes, and facilitate future rational AAV design aimed to mitigate the clinical limitations of the current technology, both reverse and forward genetic studies on AAV have been pursued. A wealth of naturally occurring AAV variants have been studied extensively and yielded large descriptive data sets (Gao et al., 2003; Gao et al., 2004; Gao et al., 2005). Forward genetic studies have resulted in important findings (Adachi et al., 2014; Asuri et al., 2012; Bartel et al., 2012; Dalkara et al., 2013; Huttner et al., 2003; Koerber et al., 2008; Li et al., 2008; Lisowski et al., 2014; Maheshri et al., 2006; Mays et al., 2013; Perabo et al., 2003; Shen et al., 2007; Wu et al., 2000) but often remain constrained by the limited tolerance for structural change of the rigid architecture of AAV (Adachi et al., 2014; Huttner et al., 2003; Lochrie et al., 2006; Mays et al., 2013; Shen et al., 2007; Vandenberghe et al., 2009; Wu et al., 2000). Rational AAV design has therefore been difficult in structurally isolating and modulating vector phenotypes while retaining integrity and the desirable aspects of its innate function and biology of the particle.
Here, we hypothesized that the divergent AAV phenotypes and structural determinants underlying its biology and pharmacology could be mapped by recreating the evolutionary lineage of this virus. Under selective pressure, evolutionary intermediates along the lineage undergo phenotypic modulation while balancing these changes with the retention of structural and functional integrity that are expected to highlight evolutionary couplings and epistatic interactions. We used ancestral sequence reconstruction (ASR) methods to predict the amino-acid sequence of putative ancestral AAV capsid monomers using maximum-likelihood (ML) methods (Finnigan et al., 2012). This led to the reconstruction of 9 nodes along the phylogeny of common ancestry of most of the AAV serotypes under clinical testing or consideration (AAV1–3 and 6–9). Through de novo gene synthesis of these reverse-translated ancestral capsid proteins, we demonstrate assembly and infectivity of members across this AAV lineage. The most distal presumed ancestor of the lineage, Anc80, was extensively characterized for its performance as a gene therapy vector and was demonstrated to be a potent broadly applicable vehicle for gene therapy with unique properties.
RESULTS
In silico ancestral sequence reconstruction of AAV capsid protein
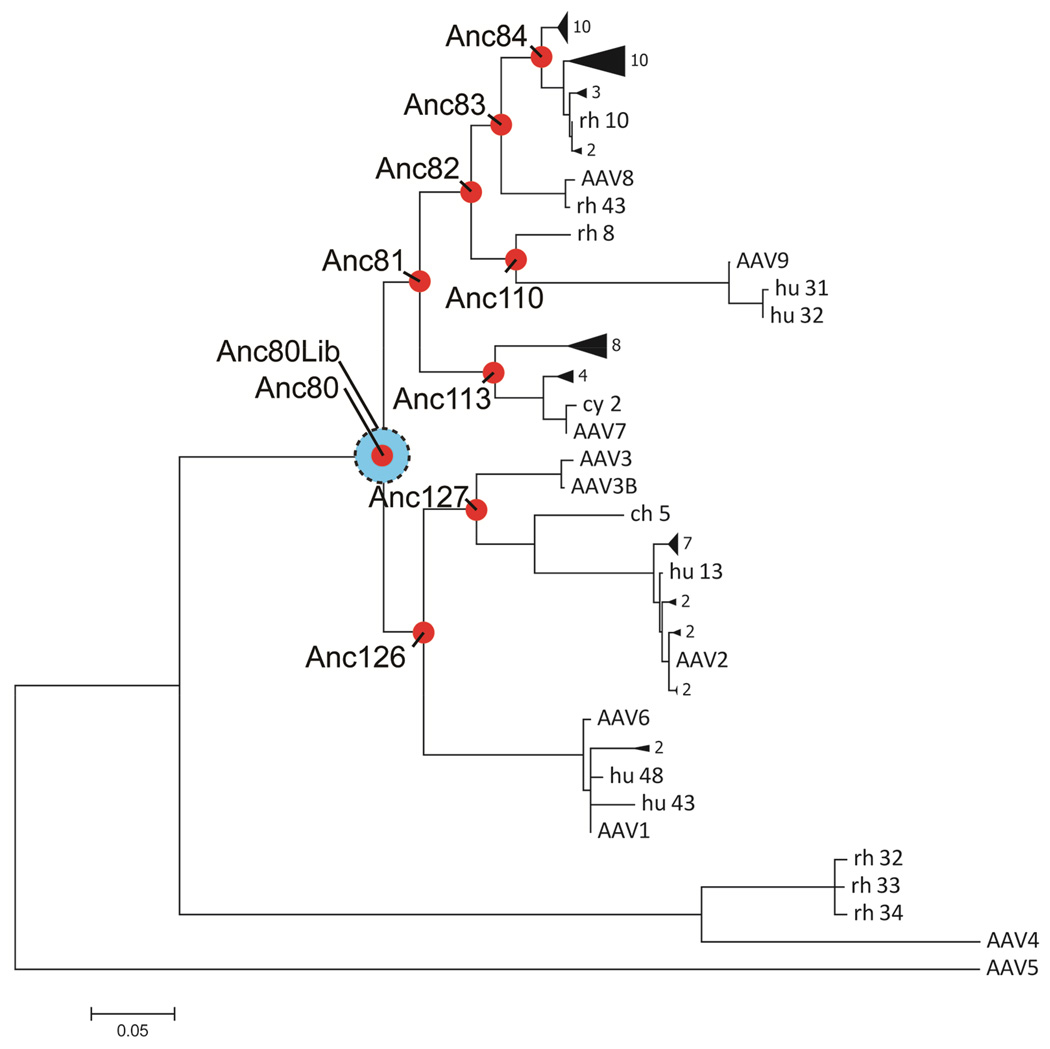
In lieu of attempting to isolate an intact ancestral viral sequence from proviral DNA or archeological samples, contemporary AAV sequence data was integrated through phylogenetic analysis and ML-ASR in order to infer the putative ancestral amino-acid sequence for the AAV Cap. A total of 75 sequences AAV serotype isolates and variants from previous biomining efforts (Gao et al., 2003; Gao et al., 2004) led to a robust AAV Cap phylogeny generated with PHYML (Guindon et al., 2010) with AAV5 as an outgroup. Only full length AAV capsids were included in this analysis that were (a) naturally occurring in primate populations, (b) previously demonstrated to assemble and infect efficiently, and (c) not known to have arisen through recombination events in its natural history, as traditional phylogenic analysis and ASR do not account for horizontal evolutionary events. The dendrogram in Figure 1 models the evolutionary path of AAV with early speciation of AAV4, and 5 serotypes, parallel to a single node, named Anc80, from which most known contemporary AAVs evolved. These serotypes include AAV1, 2, 8 and 9, currently in human gene therapy trials. Nodes in this phylogeny were named Anc and numbered sequentially. To validate our approach, the Anc80 node was developed into a recombinant virus for possible use as a gene therapy vector (Figure 1).
Figure 1. Phylogeny and ASR of AAV evolutionary lineage.
Maximum-likelihood phylogeny relating 75 isolates of AAV). Red circles represent evolutionary intermediates reconstructed through ASR. Blue circle represents library of probabilistic sequence space around Anc80. Subclades are collapsed for clarity. Full phylogeny and sequence alignment is presented in Supplementary Figure S3 and S2.
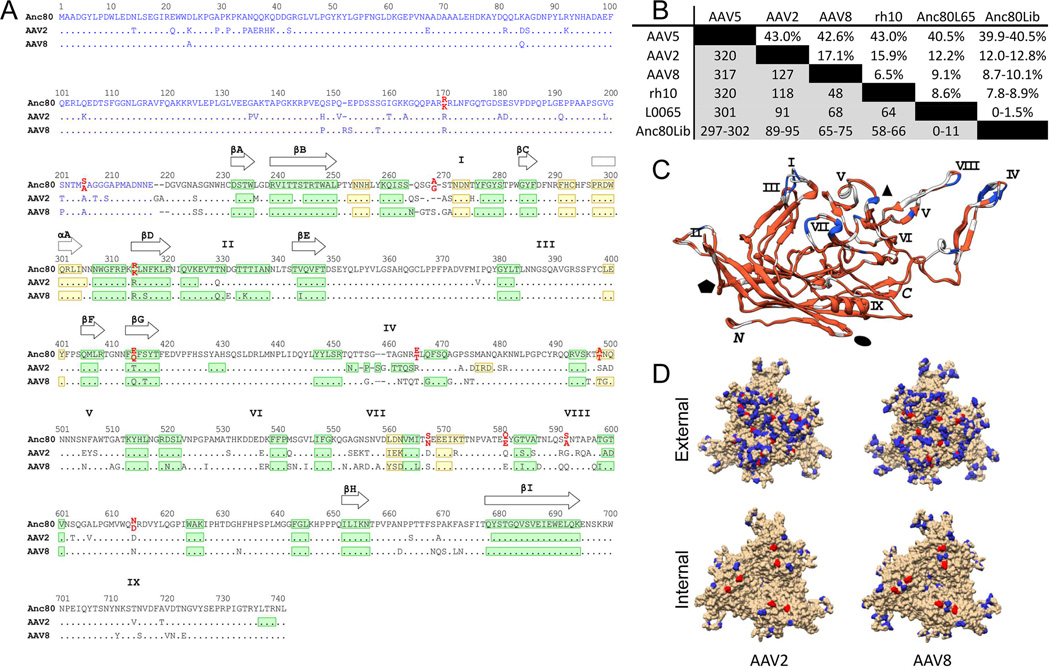
Anc80 was chosen in part because the reconstruction of this node was highly informed by the abundance of naturally occurring and clinically relevant AAV descendants. Furthermore, Anc80 is embedded in the phylogeny of the Dependoparvoviridae with known helper-dependent primate AAVs that arose prior to Anc80’s speciation (Figure 1) making it more likely that the ancestrally reconstructed particle retains the basic properties shared within this family. Using ML methods, a protein sequence prediction was derived for Anc80 based on calculated posterior probabilities for each residue in a particular position. In order to account for the uncertainty in selecting the appropriate amino-acid in each position, we aimed at generating all possible sequence permutations for positions with individual amino-acid posterior probabilities with p≥0.3. A representation of this library, Anc80Lib, is illustrated in Figure 2A in a part-structural alignment with an AAV2 and AAV8 reference capsid sequence. Practically, this led to a probabilistic sequence space composed out of sequences for which most of the 736 Anc80 capsid amino-acid positions were fixed due to the high certainty in those positions by the ML-ASR, while for 11 positions 2 amino-acid options were provided, resulting in a sequence space encompassing 211=2048 permutations.
Figure 2. Sequence and structural analysis of Anc80 vectors.
A. Sequence alignment of Anc80, AAV2 and AAV8 VP3 proteins. A structural alignment derived from the crystal structures of AAV2 (PDB 1LP3) and AAV8 (PDB 2QA0) VP3 and the predicted structure of Anc80L65 VP3 was generated with UCSF Chimera (Pettersen et al., 2004) and is represented in black print. The blue region is a non-structural alignment of the VP1/VP2 domains of AAV2, AAV8 and An80 (Notredame et al., 2000). Ambiguous residues in Anc80Lib are in red print with the lower position corresponding to Anc80L65 residues. β-strands and α-helices are represented in green and yellow, respectively. The positions of the nine β-strands forming the AAV antiparallel β-barrel are depicted with plain arrows whereas the position of the conserved core α-helix is depicted with a dotted arrow. The approximate positions of variable regions (VR) I-IX are represented by the roman numerals above the sequence alignment. B. AAV Cap Sequence divergence matrix: Table represents above the diagonal the percent sequence divergence from selected AAV serotypes, as well as rh.10, most homologous VP1 sequence as by BLAST. Below the diagonal, the number of amino-acid differences per position is presented. C. Superimposition of AAV2 and AAV8 VP3 crystal structures with Anc80L65 VP3 predicted structure. The color code depicts the amino acid conservation between the 3 aligned sequences of panel A (red: highest conservation; blue: lowest conservation). Variables regions I-IX and C/M-termini are indicated in black. The approximate positions of the two, three and five-fold axis are represented by the black ellipse, triangle and pentagon, respectively. D. Structural mapping of amino-acid changes as compared to AAV2 (left) and AAV8 (right) on VP1 trimer visualizing the external (top) and internal (bottom) of the virion. Colored residues are divergent in Anc80. Red colored residues are ambiguous via ASR and therefore dimorphic in Anc80Lib.
Structural and sequence alignment of Anc80Lib with extant AAVs and their X-ray crystallography data highlight significant divergence from currently known circulating AAV. The closest homologue as determined via BLAST search is rh.10, a rhesus macaque isolate within Clade E of the primate Dependoparvoviridae, which differs from Anc80Lib by minimally 7.8% which accounts for 58 divergent amino-acid positions (Figure 2B). AAV8 and AAV2 differ 8.7% and 12.0%, respectively and those 65–89 variable sites are distributed unevenly over the entire VP1 protein, including the VP1, 2 unique domains (Figure 2A, B). Divergence is highest in the hypervariable domains I, IV, VII, and VIII, both in terms of sequence as well as based on structural modeling of Anc80Lib clones in overlay with AAV2 and 8 monomeric structures (Figure 2A, C). Mapping of the variable Anc80 residues onto trimeric X-ray crystallography models of AAV2 and AAV8 in Figure 2D highlight most changes to be concentrated on the external surface of the virion, particularly on peak and flanks of the protrusions around the 3-fold axis of symmetry. However, a significant number of variable residues were also noted on the surface exposed domains outside of the 3-fold axis in addition to a smaller number of variations on the internal surface of the particle and on regions of Cap that are not resolved in the X ray structures.
Anc80 synthesis and basic characterization
Anc80Lib protein sequences were subsequently reverse translated and generated by gene synthesis in pooled library format. Capsid genes were cloned into an AAV packaging plasmid encoding AAV2 Rep into pAnc80Lib following which the library was deconvoluted clonally. Individual clones (named pAnc80LX, with X a consecutive number) were evaluated in isolation to avoid potentially interfering competitive interactions in a minimally divergent library population. A portion of individual Anc80 clones were Sanger sequenced verifying integrity and complexity requirements. Clonal Anc80 plasmids were co-transfected with a ΔF6 adenoviral helper plasmid, an expression construct for AAP derived from AAV2 (AAP2), and ITR flanked expression construct encoding luciferase. A total of 776 library clones were produced and inoculated at equal volume of producer cell lysate on HEK293 cells in a semi-high-throughput assay aiming to assess combined particle assembly and transduction efficiency. Approximately 50% of the Anc80 clones led to detectable signal over background in this rudimentary screening assay. Several lead candidates with highest luciferase signal progressed to sequencing confirmation and titration for DNaseI resistant genome containing particles (GC) and infectivity on HEK293 cells. Based on these results Anc80L65, the 65th Anc80Lib clone that was evaluated, was selected for further characterization. Anc80L65 vector yields from cell lysate are between 82–167% of AAV2 yields, yet were depressed compared to the high yielding AAV8 (3–5% relative AAV8 yields). In vitro infectivity on HEK293 is inferior to AAV2 however superior to AAV8 on a particle per cell basis.
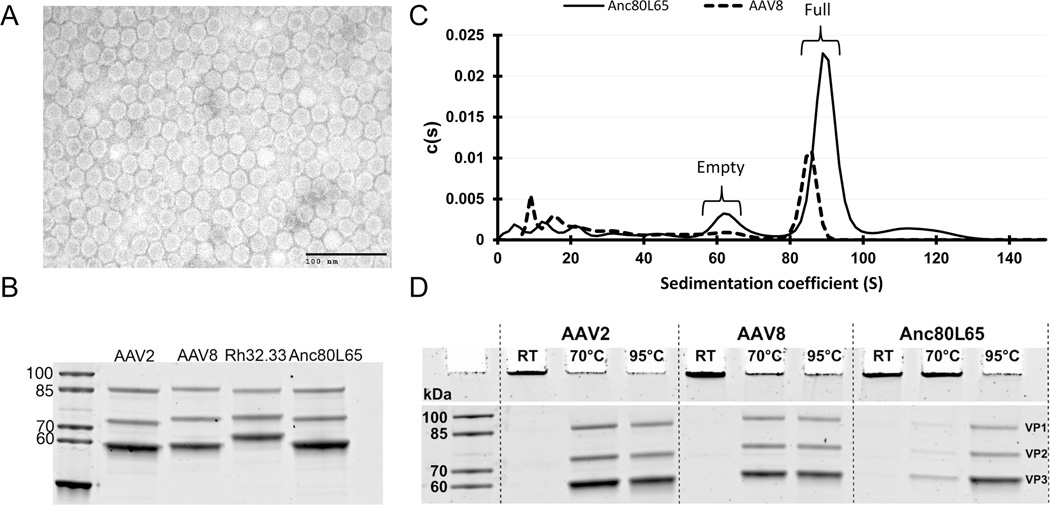
Anc80L65 vector preparations were produced and purified on an iodixanol gradient at scale following traditional protocols and subjected to a variety of biochemical, biophysical, and structural analyses. Particles within a purified preparation of Anc80L65 were visualized under negative staining by electron microscopy (EM) (Figure 3A). Anc80L65 virions present as relatively uniform hexagonally shaped particles with a diameter of approximately 20–25 nm, not unlike other AAV capsomers. Denatured particles resolved under SDS electrophoresis into 3 bands of 60, 72, and 90 kDa, in an approximate ratio of 1:1:10 corresponding to the VP1–3 proteins from AAV2 and AAV8 particles (Figure 3B). Analytical ultracentrifugation (AUC) allowed us to determine the sedimentation coefficient of genome containing Anc80L65 at 88.9 S, slightly increased from AAV8’s (85.9 S) (Figure 3C). This analysis permitted us further to determine the relative abundance of empty or lower density assembled particles, presumed to be lacking a vector genome, as well as overall purity. One concern was that inaccurate modeling of the ancestral capsid sequence may have resulted in a structure deficient in its ability to package genomes and would result in a skewed empty versus full ratio in Anc80L65 preparations. Results indicated approximately 16% empty versus 85% full particles in our preparation, in line with observations with AAV8 (Figure 3C). Additionally, we hypothesized particle stability may be reduced due to suboptimal modeling of the ancestral capsid composition, and subjected the particle to heat stability assays which determined against our expectations Anc80L65 to be 15–30° C more heat stable that its presumed A AV2 and AAV8 (Figure 3D).
Figure 3. Biophysical and biochemical characterization of Anc80L65.
A. Negative staining Transmission Electron Microscopy (TEM) of Anc80L65 particles. B. Anc80L65 viral protein composition by SDS PAGE: Purified AAV preparations were analyzed by SDS-PAGE demonstrating similar incorporation levels of monomers VP1, 2, and 3. C. Anc80L65 sedimentation and empty:full particle profile: Sedimentation coefficient distributions were derived from the refractive index optical measurement systems during analytical ultracentrifugation of AAV8 and Anc80L65 iodixanol purified preparations. D. Anc80L65 thermostability: SDS page thermostability assessment of AAV particle illustrates dissociation into monomers for AAV2 and 8 at temperatures below 70°C for AAV2 and 8 but requirin g 95°C melting temperatures for Anc80L65.
In vivo gene transfer and transduction of Anc80L65 in murine model
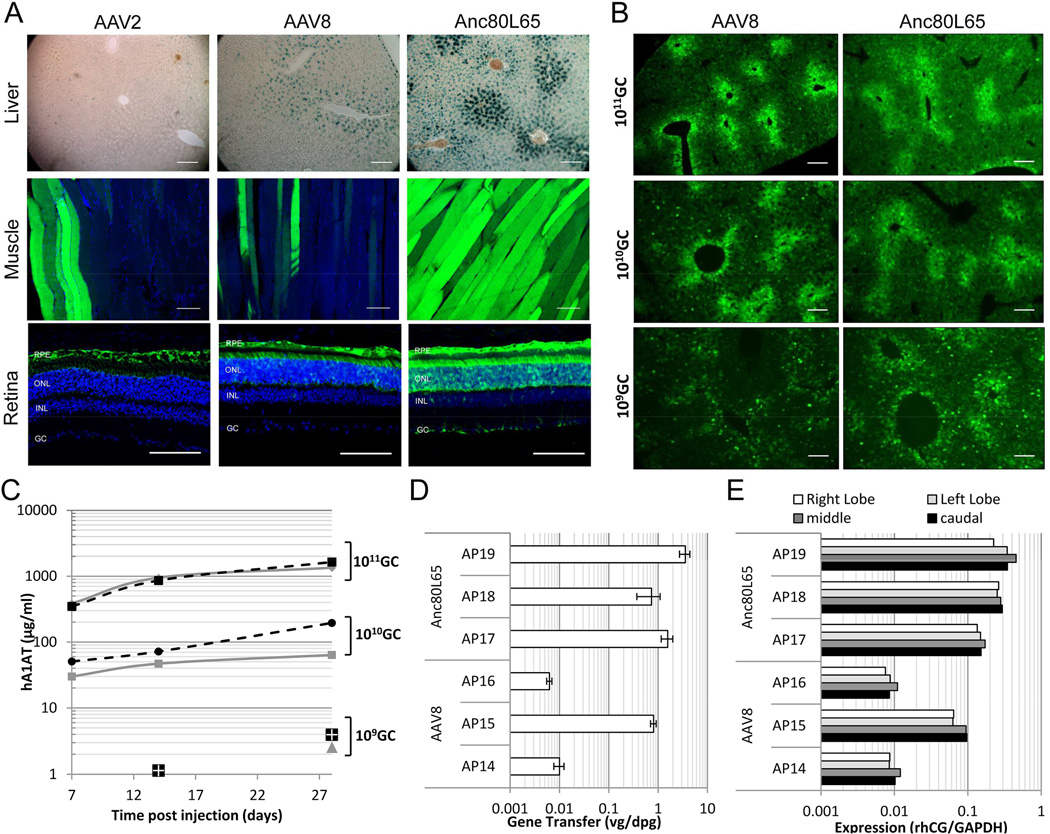
Next, we evaluated the ability of Anc80L65 packaged transgenes to be delivered and expressed from 3 clinically relevant target tissues and routes of administration (ROA) in the C57Bl/6 mouse: (a) liver following a systemic injection, (b) skeletal muscle following direct intramuscular injection, and (c) a subretinal injection for outer retina targeting. Large scale preparations of Anc80L65 were produced alongside with AAV2 and AAV8 controls with reporter genes and were injected at equal doses for liver, muscle and retina directed gene transfer in adult male C57Bl/6 mice. Expression, presented in Figure 4, was monitored qualitatively (eGFP and/or LacZ for all three target tissues and quantitatively via serum ELISA measurement of the secreted hA1AT (liver) at various time points. Liver directed gene transfer was observed to be robust via 2 routes of administration and transgenes (Figure 4A, B, C). Analogously to AAV8, hepatocytes were targeted efficiently as observed by LacZ and GFP staining surpassing the limited permissively described for AAV2 (Nakai et al., 2005; Nakai et al., 2002). Quantitatively, Anc80 demonstrated similar efficiency of transduction to AAV8 by intracellular reporter and a secreted serum protein transgene product. Dose ranging studies demonstrated a linearity of gene transfer with dose above 1010 GC/mouse but a threshold below which linearity was not maintained for hA1AT (and less obvious by eGFP) (Figure 4B, C). A biodistribution study at the high dose of 5×1011 GC/mouse was conducted at day 7 and 28 post injection to evaluate tissue distribution of vector genomes in liver, heart, spleen, kidney, and lung of Anc80L65 alongside AAV8 as a control (Table S1). Results show similar ranges of gene transfer of Anc80 to AAV8 in the tissues tested, with moderate increases for Anc80L65 in spleen, heart, and lung. Via direct skeletal intramuscular injection, Anc80 efficiently targeted myofibers proximal to the injection site and longitudinally extending across the fiber (Figure 4A and Figure S1). Retinal transduction after subretinal injection is efficient in targeting the retina pigment epithelium (RPE), as was the case in AAV2 and AAV8 as previously noted (Auricchio et al., 2001). Photoreceptor targeting, a more difficult cell target, as is documented for AAV2, was observed with AAV8 and Anc80L65 (Allocca et al., 2007). While both AAV8 and Anc80L65 targeted the majority of photoreceptor cells, transduction with Anc80L65 leads consistently to higher expression levels per cell. A limited number of cells in the inner retina were also observed to be GFP positive by Anc80L65 transduction (Figure 4A).
Figure 4. Anc80L65 in vivo transduction biology.
A, top panel: Mouse liver transduction and lacZ transgene expression comparison of AAV2, AAV8 and Anc80L65.TBG.nLacZ in liver 28 days after intraperitoneal delivery at a dose of 3.9 × 1010 GC (C57Bl/6, n=3). A, middle panel: AAV2, AAV8 and Anc80L65 muscle tropism in mouse 28 days following an intramuscular delivery at a dose of 1010 GC to the rear-right thigh (gastrocnemius) (n=5). See also Figure S1. A, lower panel: Comparison of eGFP transgene expression between AAV2, AAV8, and Anc80L65 in the murine retina after subretinal delivery at a dose of 2×109 GC. AAV2 shows high affinity for RPE cells while both RPE and photoreceptors are targeted using AAV8 and Anc80L65 vectors with Anc80L65 showing higher transduction efficiency compared to the AAV2 and 8 (C57Bl/6, n=4 eyes). B. Qualitative dose response hepatic eGFP-expression analysis following dosing of 1011 (top panel), 1010 (middle panel), and 109 (bottom panel) GC comparing AAV-8 and Anc80L65 by retro-orbital intravenous injection in mouse. Both AAV8 and Anc80L65 lead to comparable eGFP expression at equal dose throughout the dose ranging. (C57Bl/6, n=3) C. Quantitative AAV dose response analysis measuring mouse serum levels of recombinant human alpha 1-antitrypsin (hA1AT) transgene expression from AAV-8 (black symbols: square-1011 GC, circle-1010 GC, and four-square-109 GC) and Anc80L65 (grey symbols: diamond-1011 GC, square-1010 GC, and triangle-109 GC). (C57Bl/6, n=5). See also Table S1, S3, S4 and S5. D. Rhesus macaque liver gene transfer of AAV-8 and Anc80L65 expressing Rhesus chorionic-gonadotropin (rhCG) following saphenous vein injection of a dose of 1 × 1012 GC/kg. Genomic DNA was harvested from macaque liver-lobes and viral genome (vg) per diploid genome (dpg) was measured by qPCR assay. One AAV8 and all three Anc80L65 animals successfully received ~1–3 vg per diploid cell of the caudal liver lobe, while 2 AAV8 animals likely had low level NAB resulting in vector neutralization and limited liver gene transfer. See also Table S2, S6 and S7. E. Transgene mRNA expression of AAV8 and Anc80L65 in NHP caudal, right, left and middle liver-lobes by TaqMan probe-specific, quantitative reverse-transcriptase PCR (qRT-PCR). Protein levels are not available due to lacking antibody and rhCG standards. Quantitation of rhCG transcript is normalized with endogenous GAPDH mRNA levels.
Anc80L65 gene transfer and expression in non-human primate liver
Given the robust hepatotropism of Anc80L65 in mice, we aimed at evaluating gene transfer of Anc80L65 in a large animal model. Six female rhesus macaques that were previously enrolled in prior studies unrelated to AAV were injected via saphenous vein with either AAV8 or Anc80L65 at a clinically relevant dose of 1012 GC/kg (Table S2). AAVs expressing the rhesus cDNA for the β subunit of the chorionic gonadotropin (rhCG), a transgene product that the animals are tolarized for in order to avoid a non-self transgene immune response, was used as transgene. Animals were selected based on NAB to AAV8 and Anc80L65 with serum levels prior to injection below 1/4 titer. Gene transfer was assessed by Taqman qPCR for vg of total liver DNA (caudal lobe) 70–71 days following injection (Figure 4D). Surprisingly, 2 out of 3 control AAV8 injected animal had underwhelming gene transfer (<0.1 vg/dg) likely due to low level NAB at the time of injection undetectable by standard NAB assays as reported in previous studies. One AAV8 animal, presumably with no or minimal NAB to AAV8 demonstrated gene transfer levels for liver within the expected range of 0.81 vg/dg. Anc80L65 gene transfer was efficient with 3 animals yielding hepatic transgene copy numbers ranging from 0.73–3.56 vg/dg. Liver expression was monitored via quantitative RT-PCR (Figure 4E): Anc80L65 led to expression superior to the AAV8, and achieved rhCG transcript levels between 13–26% of total GAPDH mRNA amounts in all liver lobes.
Safety, Immunology, and Toxicology of Anc80L65
The consideration to use any efficient gene delivery vector system for therapeutic application requires extensive evaluation of its safety for clinical use. In addition, the use of a novel agent which may approximate an ancestral state of a Dependoparvovirus may further raise those concerns. Here, in a non-formal preclinical setting, we examined several important aspects that may limit Anc80L65 from a safety perspective. Animal expression studies (Fig. 4) were monitored for obvious signs of toxicity during the in life phase of the study and for target tissue specific toxicity. No notable adversity was found to be associated with the vector injection. Briefly, vector administration following intraperitoneal (maximum dose tested [mdt]: 3.9 × 1010 GC/mouse), retro-orbital vein injection (mdt: 5 × 1011 GC/mouse), subretinal (mdt: 2 × 109 GC/eye), intravitreal (mdt: 2 × 109 GC/eye), and direct intramuscular (mdt: 1010 GC/mouse) were not observed to have overt toxicity. A more direct assessment was performed in a high dose intravenous injection of 5 × 1011 GC/mouse (equivalent to approximately 2 × 1013 GC/kg) of Anc80L65.TBG.eGFP alongside the following controls: (a) AAV8 with the same transgene cassette, and (b) an equal volume saline injection. Mice were phlebotomized pre-injection, 2 hours (h), 1, 3, 7, 14, and 28 days (d) post injection and blood was analyzed for Cell Blood Counts (CBC) and Serum Chemistry (Chem) (Tables S3, S4). CBC/Chem values for Anc80L65 were within normal range or comparable to controls. Serum from the 2 h, 24 h, 3 d, and 7 d time points were further evaluated for cytokines as a measure of innate immune response to the vector antigens by multiplex 23 cytokine analysis (Table S5). Cytokines for Anc80L65 were overall concordant with those for saline and AAV8 control serum, and no major cytokine elevations or decreases were observed, however in some instances were moderately outside the ranges set by the saline control values in a manner that was more apparent for Anc80L65 than AAV8. Similar analyses were performed on the blood from the rhesus studies described in Figure 4 D and E. Analogous to the mouse studies, CBC and Chem values did not demonstrate e signs of toxicity related to the AAV8 or Anc80L65 test article (Tables S6, S7)
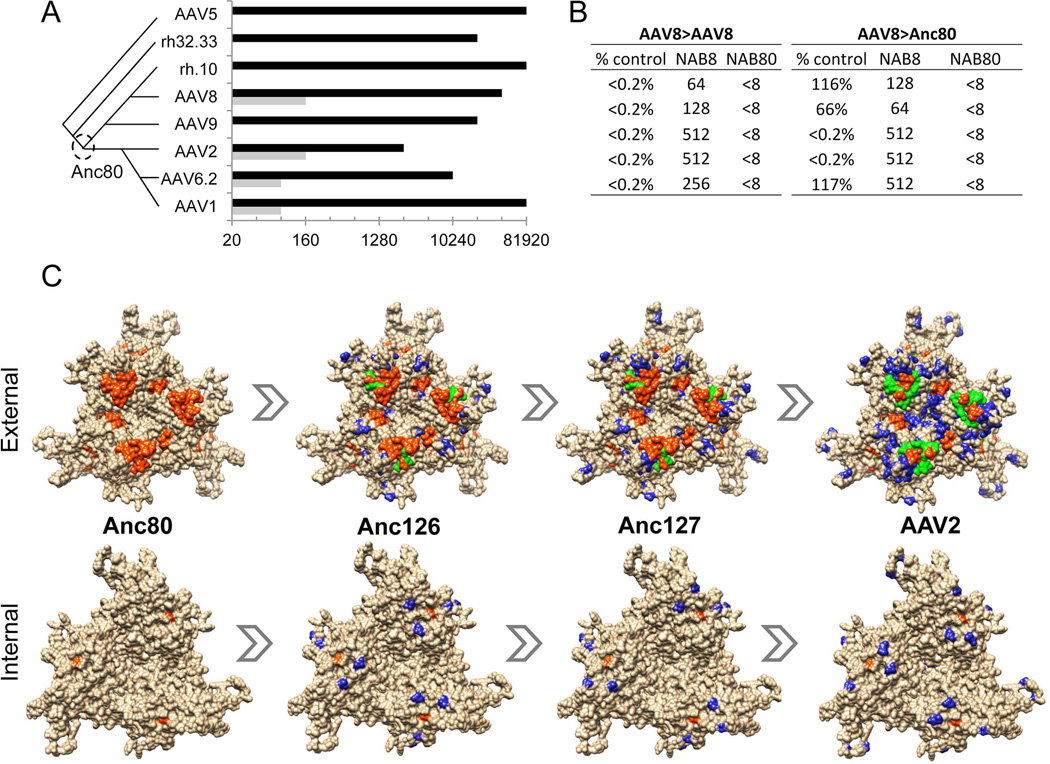
Pre-existing immunity to AAV serotypes is known to block gene transfer, and may put the patient at risk for adversity due to recall of memory T-cells toward vector antigens shared with the naturally occurring wild type virus involved in the primary infection. We used high titer rabbit antiserum raised against AAV serotypes 1, 2, 5, 6.2, 8, 9, and rh.32.33. We furthermore included rh.10 as its sequence is most closely homologous to Anc80L65 differing in 8.6% of residues. In Figure 5A, sera were tested for their ability to neutralize Anc80L65 versus the homologous vector capsid it was raised against. Results demonstrate no cross-reactivity to the structurally highly divergent AAV5 and rh32.33, while AAV2, 6.2, and 8, presumed descendants of Anc80L65 demonstrated low level cross-reactivity with between 16–1024 fold reduced neutralization as compared to the homologous virus. Among Anc80 lineage members, no cross-reactivity was observed above the limit of sensitivity for AAV9 and rh.10. Next, we aimed at validating these results in an in vivo model for neutralization by pre-immunizing animals for AAV8 via intramuscular route, and 25 days following the immunization assessing the neutralization of Anc80L65 following intravenous injection in comparison to AAV8 (Figure 5B). Neutralization was complete for AAV8 in the AAV8 pre-immunized animals. Anc80L65 was neutralized in 2/5 animals, yet demonstrated between 60–117% of transduction in 3/5 animals notwithstanding demonstrated AAV8 NAB in those animals. These results demonstrate partial cross-reactivity of Anc80L65 with AAV8 in rabbit and mouse.
Figure 5. Immunological characterization of Anc80L65.
A. Rabbit anti-AAV serum cross-reactivity: Rabbit antiserum raised against AAV serotypes (Y-axis) was tested for NAB to Anc80L65 (grey bars) versus the homologous AAV serotype (black bars) in order to assess relative sero-cross-reactivity. Values (X-axis) represent highest dilution at which >50% neutralization is achieved. Phylogenetic relationship between immunizing serotypes is depicted schematically on the left. B. Mouse in vivo gene transfer cross-neutralization: C57Bl/6 mice received an IV injection of AAV8 or Anc80L65.CASI.EGFP.2A.A1AT 25 days following an IM injection with either saline or AAV8.TBG.nLacZ. 14 days following the second injections serum was titrated by ELISA for hA1AT expression. Table presents the relative hA1AT levels of the pre-immunized mice versus the non-immunized for each vector (% control), and the NAB titer dilutions for AAV8 (NAB8) and Anc80L65 (NAB80) 24 h prior to the second injection in the immunized group (n=5). C. A non-structural multiple sequence alignment between Anc80, Anc126, Anc127 and AAV2 VP3 sequences was generated using the Tcoffee alignment package. AAV2 trimer structure was generated using UCSF Chimera. The blue residues represent residues different from Anc80. The orange residues are defined T and B-cell epitopes on AAV2 (Gurda et al., 2013; Mingozzi et al., 2007). Green residues show the overlap between orange and blue residues to highlight mapped epitopes altered in the putative evolutionary intermediates. Human T-cell epitopes with MHC haplotype: VPQYGYLTL (B*0702), SADNNNSEY (A*0101), YHLNGRDSL (B*1501), and TTSTRTWAL (B*0801). (Mingozzi et al., 2007) Mouse B-cell epitopes of defined AAV2 antibody SADNNNS plus RGNRQ for C37B Fab (Gurda et al., 2013). In bold are the residues within each epitope that are distinct between Anc80L65 and AAV80.
AAV Lineage analysis and reconstruction
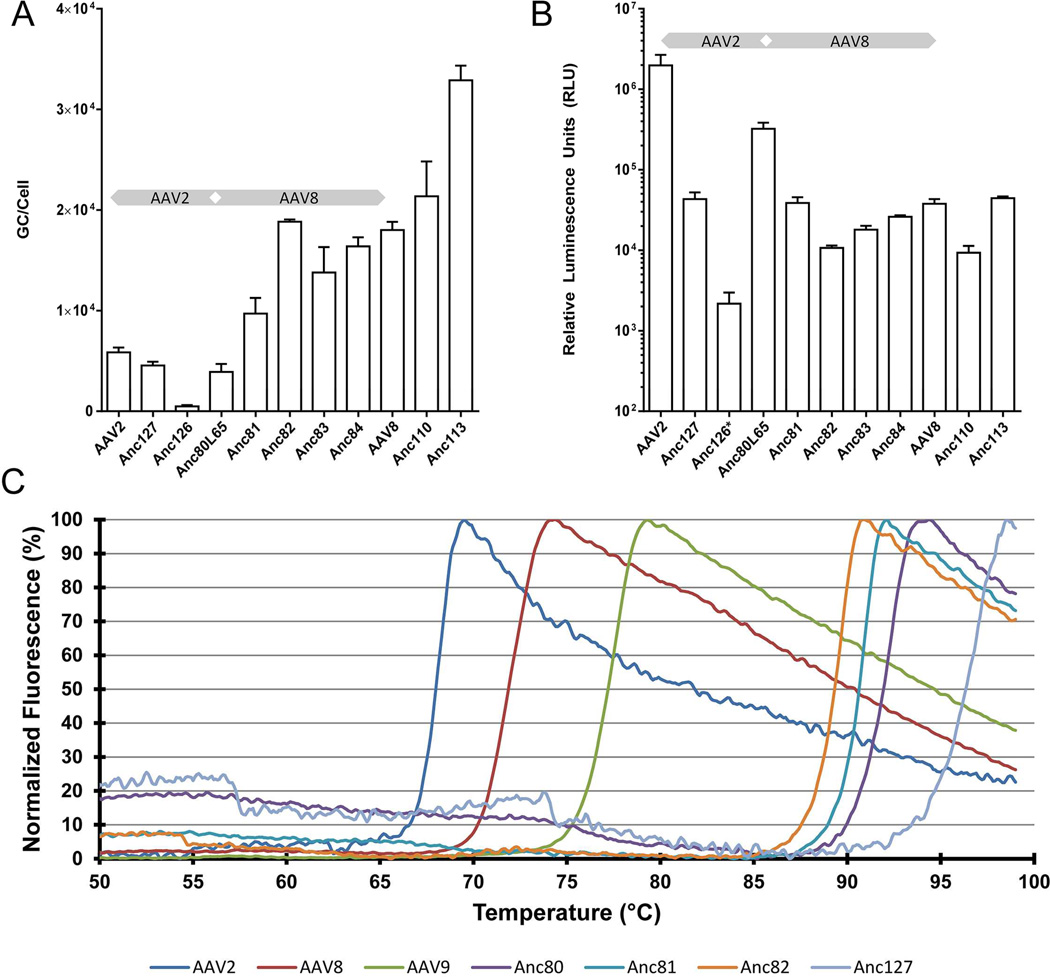
Strengthened by the successful synthesis of Anc80L65 based on ASR and its demonstration as producible, stable, and highly infectious agent for gene therapy, we aimed at providing additional validation of our approach and modeling methodology by reconstructing the lineage of AAV further. Our ambition generating this additional set of reagents was to provide structural intermediates of phenotypically distinct AAVs permitting empirical evaluation of the structure-function relationship within this viral family and highlighting important epistatic couplings informative to future AAV rational design approaches. A total of 8 additional evolutionary intermediates of AAV were reconstructed by ASR and synthesized in the laboratory (Figure 1): Anc81, Anc82, Anc83, Anc84, Anc110, and Anc113 were resolved in the branching leading toward AAV7, 8, and/or 9, while Anc126 and Anc127 are positioned in the natural history of AAV1, 2, and/or 3. For each of these, the sequence was determined by selecting the amino-acid with highest posterior probability per position (Fig. S2). First, we determined GC viral vector yields following HEK293 standard triple transfection by Taqman qPCR. Results, shown in Figure 6A, demonstrate increased productivity from Anc80 as the putative ancestor in the AAV7–9 lineage, in line with the higher production yields of those serotypes such as AAV8. The AAV1–3 branch did not present yield increases, and a very poor particle yield for Anc126. It is possible that Anc126 yields can be improved upon through leveraging the statistical space as was the case for Anc80. It is equally however likely that Anc126 ASR is less informed due to under-sampling of this branch of the AAV phylogeny. We further tested infectivity of the produced particles at equal particle doses in vitro on HEK293 by GFP and luciferase. All newly synthesized Anc vectors demonstrated infectivity, however at varying degrees (Figure 6B). In the AAV7, 8 & 9 lineage infectious titers were overall depressed and more similar to the AAV8 phenotype than that of Anc80. Anc127, the only intermediate in the Anc80 to AAV2 lineage that could be tested at equal dose demonstrated declined transduction efficiency as compared to both Anc80 as well as AAV2. We further tested the heat stability profile of selected evolutionary intermediates in both branches of this lineage (Figure 6C). Interestingly Anc81 and Anc82 demonstrated high, yet moderately decreased melting temperature in a thermostability assay compared to Anc80L65, suggesting a gradual reduction of particle denaturation temperature with evolutionary age in this branch. In contrast, Anc127 demonstrated an even further increase from the already highly thermostable Anc80L65 vector.
Figure 6. AAV lineage reconstruction modulates production, infectivity, and thermostability.
A. Production of nine ancestral and two extant viral vectors containing a luciferase reporter gene driven by a CMV promoter determined by qPCR. Error bars represent standard deviation of three biological replicates. B. Ancestral and extant viral vectors were used to transduce HEK293 cells at a particle to cell ratio of 1.9 × 103. Error bars represent standard deviation of three distinct lots of vector. *Anc126, was added at ratios between 2.1 × 102 and 3.5 × 102 GC/cell due to low vector yield. Grey diverging arrow in A and B panels schematically illustrate AAV2 and AAV8 lineage phenotypic evolution. C. Sypro orange thermostability assay indicating denaturation temperatures of selected ncestral and extant AAV vectors. See also Figure S2.
Lastly, we explored the ability of ASR to disrupt known epitopes to AAV2. Only few B or T-cell epitopes have been mapped on AAV2 to date, all of which were mapped onto Anc80L65, Anc126, Anc127, and AAV2, representing the AAV2 lineage. The introduction of the sequential mutations between these putative evolutionary intermediates highlights in Figure 5C the overlap between the mutations and 2/4 human T-cell epitopes (Mingozzi et al., 2007), and 2/2 mouse B-cell epitopes (Gurda et al., 2012). These data highlight the potential of ASR to be used as a method to eliminate or modulate antigenic regions onto the AAV capsid, and may suggest immunity was a major selective pressure in the natural history of AAV.
DISCUSSION
Forward genetics studies on multimeric complex protein assemblies are intricately difficult due to the structural constraints imposed by secondary, tertiary, and quaternary interactions within and between monomers. Many virus form icosahedral capsids as an economical way to build larger complex shells from smaller repeat units without expending excessive genetic bandwidth. These 20-faceted structures are composed of 60, or multiples thereof, subunits, each of which can integrate one or more structural viral proteins. As a function of its triangulation number and monomer size, Parvoviridae, with AAV as a member, are therefore one of the smallest known virus with a capsid diameter in the 18–26 nm range (Knipe and Howley, 2013). In order for the virus to evolutionary maintain the benefits from this icosahedral structure, sequence permutations are iteratively evaluated, not unlike traditional forward genetic structure-function experimentation in a laboratory setting. Only those monomer conformations that efficiently integrate in the icosahedral structure and can provide a selective advantage are able to persist, ultimately driving speciation.
Here, we sought to investigate, reconstruct, and learn from this natural experimentation using the simplest of all icosahedral viruses, AAV, as a model. Here, we aimed at reconstructing the viral lineage of AAV, a commonly used virus for gene transfer through methodologies similar to Gullberg et al.’s reconstruction of Coxsackievirus putative ancestor (Gullberg et al., 2010). First, putative ancestral sequences of the AAV capsid protein was inferred using in silico phylogenetic and statistical modeling. Next, these sequences were synthesized de novo and using traditional virological technique the virions derived were evaluated for assembly and infectivity. We anticipated the solution space containing monomers that structurally integrate into a viral particle and result in infectious virions to be minimally small, given that the limited knowledge base on intra- and intermolecular epistatis or the minimal structural components of AAV that constitute infectivity or function. To hedge for this anticipated low probability of success, ML statistics were used to inform us; we generated a probabilistic sequence space providing margins to account for the ambiguity of the heuristics used. Screening of the vector library that emerged from this sequence space yielded Anc80L65: a viral-like, AAV-like particle via structural and biochemical measures (Figure 2, 3) and 8.6% or 64 AA divergent from the closest known AAV sequence. Anc80L65 furthermore demonstrates high stability, unlike any of the tested extant descendants. Anc80L65 is infectious (Fig. 6B), and is efficient as a gene transfer vehicle in murine models for retina, liver, and muscle targeting and in non-human primates for liver directed gene transfer (Fig. 4). Its utility as a gene transfer vector was further evaluated in a set of safety studies which did not illustrated vector-related toxicity concerns (Tables S1–7).
An important property of a gene transfer vector for clinical utility is its ability to be produced efficiently. Vector yields are a function of the production system and innate structural assembly efficiencies. Anc80L65, exceeding our expectation, yields GC particles in approximately equivalent ranges to AAV2, the most studied clinical AAV technology to date. AAV2 yet is reduced compared to other high yielding AAVs (Fig. 6A). In an attempt to understand the variation in particle yield, we tested the hypothesis that this is due to reduced stability. In contrast, Anc80L65 was thermostable up to temperatures of 92° C compared to 68° and 72° C for AAV2 and 8. It is possible that this high thermostability informs us on a higher activation energy threshold needed for Anc80L65 to dissociate, and possibly inversely, to assemble. Further work is required to investigate this fascinating relationship. Another possibility relates to what can be referred to as the ‘Back to the Future’ concern; here we present data on an attempt to take the ancestral state of a single virion component, and bring it to life in an otherwise fully contemporary context. This process conceptually ignores the possibility that coevolution took place with other AAV components (e.g. AAP, ITR), helper functions provided by adenovirus or other helper virus functions, and the host cell. If indeed such co-evolution occurred, those heterologous ancestral components may be required to bring full functionality to Anc80L65, or any ancestral AAV capsomer. To date, no data is available to suggest indeed such a missing ancestral link is prohibiting further potentiation of Anc80L65 in its assembly, packaging, or transduction biology.
Building on the validation that Anc80L65 brought to our methodology, we expanded the set of evolutionary intermediates for experimentation to 8 additional Anc particles along the lineage toward AAV1–3 and 7–9 (Figure 1). We demonstrate assembly, packaging, and in vitro infectivity of these particles. Particularly along the AAV8 branching from Anc80L65, our data suggest an increased productivity of viral yields illustrating that close putative descendants of Anc80L65 were able to overcome the production limitation discussed above (Figure 6).
In addition to ASR providing a novel methodology to synthetically derive viral vectors with novel biology, as demonstrated for Anc80L65, the availability of functional intermediates of divergent extant viruses may enable the elucidation of relevant questions to the unique biology of the distinct AAV serotypes. In turn, these data may ultimately make possible structure-guided design of AAV, and tailor it to the specific requirement of a clinical application. Our approach may furthermore shed light on the evolutionary pressures of AAV along its modeled natural history. One such presumed evolutionary pressure is of highest concern for gene therapy applications. Since AAV is widely circulating in humans, long-lived memory T and B-cell responses pose a problem for gene therapy across broad populations. Indeed, individuals with pre-existing immunity (PEI) to AAV (in some populations over 50%), are currently excluded from participating in many AAV clinical trials as no adequate alternative mitigation strategies exist to address the problem (Louis Jeune et al., 2013). Here, we demonstrate that ASR is able to modulate known epitopes that govern B and T-cell PEI to novel antigenic sites and thereby possibly reduce the affinity of MHCI-TCR and virus-NAB interactions (Fig. 5). Unlike rabbit vaccination experiments as in Figure 5A, human PEI is highly cross-reactive across many serotypes, making it difficult for any AAV-like particle escape this wide breadth PEI in humans (Brantly et al., 2009; Calcedo et al., 2009). To evaluate seroprevalence of Anc vectors and how these data inform a gene therapy outcome, a more robust and predictive assay is required as are extensive seroprevalence studies. Indeed our data highlights traditional NAB assays poorly correlating with in vivo neutralization (Figure 4D and 5B), as was previously noted (Wang et al., 2010).
In conclusion, ASR is shown to be a powerful methodology to generate functional intermediates of complex and structurally constrained biological assemblies, here exemplified for an icosahedral virus, AAV. The ancestral reconstruction of Anc80L65, the common putative ancestor of AAV1–3 and 7–9, yielded a highly potent vector particle with potential use in gene therapy applications via in silico and synthetic biology methods. The resolution of the lineage from Anc80 toward these extant serotypes provides a toolset of gene transfer reagents that further can help elucidate complex structure-function relationships within AAV and eventually may facilitate structure-based design of this potential new class of genetic drugs.
EXPERIMENTAL PROCEDURES
Ancestral Sequence Reconstruction of AAV Capsids
These studies were conducted following approval of the Harvard Medical School Institutional Biosafety Committee (Committee on Microbiological Safety) which included considerations of dual use of the vector reagents described here. Ancestral capsid sequences were reconstructed using maximum-likelihood methods as in (Finnigan et al., 2012) An alignment of 75 AAV capsids (Genbank accession numbers in supplement) was generated using PRANK v.121002 using the –F option (Loytynoja and Goldman, 2005), (Loytynoja and Goldman, 2008) and the JTT+F+G model was determined to be the phylogenetic model of best fit through the Aikake Information Criterion as implemented in ProtTest3 (Darriba et al., 2011). The alignment and best-fit model were then used to infer a phylogeny through PhyML 3.0 (Guindon et al., 2010) which was evaluated through the approximate likelihood-ratio test (aLRT) (Anisimova and Gascuel, 2006) as implemented in PhyML. Ancestral capsid sequences were then inferred using PAML 4.6 (Yang, 2007) through the Lazarus package developed by the Thornton group. Sequences are available through GenBank (KT235804-KT235812). In order to compensate for the uncertainty inherent to the reconstruction, a script was written to assess the computed posterior probabilities to identify ambiguously reconstructed sites. Positions were considered ambiguous along the reconstructed capsid sequence if more than one amino-acid had a posterior probability greater than 0.3. Eleven such sites were identified on Anc80, each with two probable amino acids. These eleven dimorphic sites were then incorporated into a DNA library using the codons from a modern virus (rh.10). Because the reconstruction did not consider the coevolution of AAP and the capsid, the AAP open-reading frame was ablated by changing the non-canonical CTG start codon to CAG during library design. In addition, another downstream ATG also in the AAP ORF was ablated by changing the codon to AAG. These modifications did not alter the amino-acids in the cap ORF. The DNA library was then synthesized by DNA2.0 and subsequently subcloned into expression vectors via restriction enzyme digest and ligation.
In vitro Characterization of AAV Ancestral Lineage Vectors
To identify and characterize functional AAV capsids within Anc80Lib, individual clones from the subcloned DNA library were isolated and used to produce luciferase-containing vector in either 6-well or 96-well with AAP2 provided in trans. Crude vector was isolated by filtering cell lysate through a 0.4 µm filter after 48 hours had elapsed since transfection. Next, equal volumes of this crude vector preparation were added to 96-well plates confluent with HEK293 cells which were evaluated for their luciferase activity an additional 48 hours later. In total, 776 clones were evaluated. Crude preparations of vector containing a CMV driven luciferase were produced by triple transfection in a 6-well format, supplementing AAP in trans to ancestral AAV vectors. In total, three different independent biologic replicates were produced per vector. DNAseI resistant transgenes were quantified as above. These crude preparations of virus were each then evaluated for their ability to transduce HEK293 cells in technical triplicates at an MOI of 1.9E3 GC/cell with the exception of Anc126, which was added at MOIs between 2.1E2 and 3.5E2 gc/cell. After 48 hours had elapsed, the transduced cells were assessed for luciferase via luminescence assay.
AAV Vector Preparation
Large-scale polyethylenimine (PEI) transfections of AAV cis, AAV trans, and adenovirus helper plasmid were performed in a 10-layer hyperflask (Corning) with near confluent monolayers of HEK 293 cells. Plasmids were transfected at a ratio of 2:1:1 (260 µg of adenovirus helper plasmid/130 µg of cis plasmid/130 ug of trans plasmid). Transfections for production of Anc vectors were supplemented with pAAP2 in equivalent amounts as the AAV cis plasmid. PEI Max (Polysciences, Warrington, PA)/DNA ratio was maintained at 1.375:1 (w/w). The transfection and downstream purification process were performed as previously described (Lock et al., 2010). DNAseI-resistant vector genomes copies were used to titrate AAV preparations by TaqMan qPCR amplification (Applied Biosystems 7500, Life Technologies) with primers and probes detecting promoter, transgene, or poly-adenylation signal coding regions of the transgene cassette. The purity of the large-scale preparations was evaluated by SDS-PAGE gel electrophoresis.
In vivo gene transfer (mouse)
C57BL/6 male mice (6–8 weeks old) were purchased from Charles River Laboratories (Wilmington, MA) and kept at the Schepens Eye Research Institute (SERI) Animal Facility. All animal procedures were performed in accordance with protocols approved by the Institute of Animal Care and Use Committees at SERI. For liver directed studies, Male C57BL/6 mice were each injected retro-orbitally (150µl) or intraperitoneally (100µl). Blood was collect via submandibular bleeds using GoldenRod animal lancets (MEDIpoint, Inc.). Animals were euthanized, and livers were collected. Expression of lacZ was assessed using a β-Gal staining kit (Life Technologies, Cat. No. K1465-01). In skeletal muscle studies, male C57BL/6 mice were injected into the rear-right gastrocnemius muscle (50 uL). Retinal studies were performed by subretinal injection of male C57BL/6 mice (2 uL).
In vivo gene transfer (nonhuman primate)
Experiments with rhesus monkeys were performed at New England Primate Research Center. All experimental procedures were approved by the Office for Research Subject Protection, Harvard Medical Area (HMA) Standing Committee on Animals, the Harvard Medical School Institutional Animal Care and Use Committee. Animals were sedated with ketamine or telazol in combination with dexdomitor. AAV was administered intravenously via the saphenous vein in a 20 ml volume at an approximate rate of 1ml/min. After recovering from the injection, the animals were monitored clinically for general wellbeing and followed for 2 months. During this time phlebotomies were performed at regular intervals to evaluate immune response to AAV and toxicity. After 70 days monkeys were euthanized, and liver samples were harvested. Tissue was harvested in RNALater (Life Technologies).
Additional detail on the experimental procedures is provided in detail in the supplement.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Rajani Shelke and Yang Lin from the MEEI/SERI Gene Transfer Vector Core (http://vector.meei.harvard.edu), James M. Wilson for sharing vector plasmid and rabbit antiserum reagents, George Church, Eric Pierce, and Jason Comander for discussion, Sean Gai and Alden Harwood for technical support and computational analysis, Debalina Sarkar provided technical assistance with multiplex cytokine analysis, Debby Pheasant (Biophysical Instrumentation Facility, CSBi at MIT) for training and assistance on AUC, Phil Seifert (SERI Morphology Core). Support for this work comes from The Harvard Medical School Department of Ophthalmology, Mass Eye and Ear, the Grousbeck Family Foundation, The Candyce Henwood Fund, The NIH Common Fund (5DP1EY023177-03), Research to Prevent Blindness, and Foundation Fighting Blindness.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
EZ conceived, designed and conducted in silico and in vitro experiments and helped with the drafting of the manuscript; SP and AM conceived and performed structural modeling and thermostability studies. VK, HTT, RS, SS, RX, EP, and LSC conducted various aspects of the immunological and/or in vivo experiments. EAM and EP contributed to the manuscript and scientific management of the program. LHV conceived, supervised the scientific program, and wrote the manuscript.
CONFLICT OF INTEREST STATEMENT
LHV and EZ are inventors on a patent describing some of the methods and reagents described here. LHV is inventor on several patents on gene therapy technologies. LHV is co-founder, shareholder, member of the Scientific Advisory Board, and consultant of GenSight Biologics, an ophthalmology gene therapy company.
REFERENCES
- Adachi K, Enoki T, Kawano Y, Veraz M, Nakai H. Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nature communications. 2014;5:3075. doi: 10.1038/ncomms4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allocca M, Mussolino C, Garcia-Hoyos M, Sanges D, Iodice C, Petrillo M, Vandenberghe LH, Wilson JM, Marigo V, Surace EM, et al. Novel adeno-associated virus serotypes efficiently transduce murine photoreceptors. J Virol. 2007;81:11372–11380. doi: 10.1128/JVI.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Systematic biology. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Asuri P, Bartel MA, Vazin T, Jang JH, Wong TB, Schaffer DV. Directed evolution of adeno-associated virus for enhanced gene delivery and gene targeting in human pluripotent stem cells. Mol Ther. 2012;20:329–338. doi: 10.1038/mt.2011.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auricchio A, Kobinger G, Anand V, Hildinger M, O'Connor E, Maguire AM, Wilson JM, Bennett J. Exchange of surface proteins impacts on viral vector cellular specificity and transduction characteristics: the retina as a model. Hum Mol Genet. 2001;10:3075–3081. doi: 10.1093/hmg/10.26.3075. [DOI] [PubMed] [Google Scholar]
- Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, Viswanathan A, Holder GE, Stockman A, Tyler N, et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- Bartel MA, Weinstein JR, Schaffer DV. Directed evolution of novel adeno-associated viruses for therapeutic gene delivery. Gene therapy. 2012;19:694–700. doi: 10.1038/gt.2012.20. [DOI] [PubMed] [Google Scholar]
- Bell CL, Gurda BL, Van Vliet K, Agbandje-McKenna M, Wilson JM. Identification of the galactose binding domain of the adeno-associated virus serotype 9 capsid. J Virol. 2012;86:7326–7333. doi: 10.1128/JVI.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantly ML, Chulay JD, Wang L, Mueller C, Humphries M, Spencer LT, Rouhani F, Conlon TJ, Calcedo R, Betts MR, et al. Sustained transgene expression despite T lymphocyte responses in a clinical trial of rAAV1-AAT gene therapy. Proc Natl Acad Sci U S A. 2009;106:16363–16368. doi: 10.1073/pnas.0904514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant LM, Christopher DM, Giles AR, Hinderer C, Rodriguez JL, Smith JB, Traxler EA, Tycko J, Wojno AP, Wilson JM. Lessons learned from the clinical development and market authorization of Glybera. Hum Gene Ther Clin Dev. 2013;24:55–64. doi: 10.1089/humc.2013.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcedo R, Vandenberghe LH, Gao G, Lin J, Wilson JM. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis. 2009;199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkara D, Byrne LC, Klimczak RR, Visel M, Yin L, Merigan WH, Flannery JG, Schaffer DV. In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci Transl Med. 2013;5:189ra176. doi: 10.1126/scitranslmed.3005708. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics (Oxford, England) 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMattia MA, Nam HJ, Van Vliet K, Mitchell M, Bennett A, Gurda BL, McKenna R, Olson NH, Sinkovits RS, Potter M, et al. Structural insight into the unique properties of adeno-associated virus serotype 9. J Virol. 2012;86:6947–6958. doi: 10.1128/JVI.07232-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan GC, Hanson-Smith V, Stevens TH, Thornton JW. Evolution of increased complexity in a molecular machine. Nature. 2012;481:360–364. doi: 10.1038/nature10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Alvira MR, Somanathan S, Lu Y, Vandenberghe LH, Rux JJ, Calcedo R, Sanmiguel J, Abbas Z, Wilson JM. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6081–6086. doi: 10.1073/pnas.0937739100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, Wilson JM. Clades of Adeno-associated viruses are widely disseminated in human tissues. Journal of virology. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Vandenberghe LH, Wilson JM. New recombinant serotypes of AAV vectors. Current gene therapy. 2005;5:285–297. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- Girod A, Wobus CE, Zadori Z, Ried M, Leike K, Tijssen P, Kleinschmidt JA, Hallek M. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J Gen Virol. 2002;83:973–978. doi: 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- Grieger JC, Snowdy S, Samulski RJ. Separate basic region motifs within the adeno-associated virus capsid proteins are essential for infectivity and assembly. J Virol. 2006;80:5199–5210. doi: 10.1128/JVI.02723-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Gullberg M, Tolf C, Jonsson N, Mulders MN, Savolainen-Kopra C, Hovi T, Van Ranst M, Lemey P, Hafenstein S, Lindberg AM. Characterization of a putative ancestor of coxsackievirus B5. J Virol. 2010;84:9695–9708. doi: 10.1128/JVI.00071-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurda BL, DiMattia MA, Miller EB, Bennett A, McKenna R, Weichert WS, Nelson CD, Chen WJ, Muzyczka N, Olson NH, et al. Capsid antibodies to different adeno-associated virus serotypes bind common regions. J Virol. 2013;87:9111–9124. doi: 10.1128/JVI.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurda BL, Raupp C, Popa-Wagner R, Naumer M, Olson NH, Ng R, McKenna R, Baker TS, Kleinschmidt JA, Agbandje-McKenna M. Mapping a neutralizing epitope onto the capsid of adeno-associated virus serotype 8. J Virol. 2012;86:7739–7751. doi: 10.1128/JVI.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie E, Samulski RJ. Adeno-Associated Virus at 50: A Golden Anniversary of Discovery, Research, and Gene Therapy Success-A Personal Perspective. Hum Gene Ther. 2015 doi: 10.1089/hum.2015.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner NA, Girod A, Perabo L, Edbauer D, Kleinschmidt JA, Buning H, Hallek M. Genetic modifications of the adeno-associated virus type 2 capsid reduce the affinity and the neutralizing effects of human serum antibodies. Gene Ther. 2003;10:2139–2147. doi: 10.1038/sj.gt.3302123. [DOI] [PubMed] [Google Scholar]
- Jacobson SG, Cideciyan AV, Ratnakaram R, Heon E, Schwartz SB, Roman AJ, Peden MC, Aleman TS, Boye SL, Sumaroka A, et al. Gene therapy for leber congenital amaurosis caused by RPE65 mutations: safety and efficacy in 15 children and adults followed up to 3 years. Arch Ophthalmol. 2012;130:9–24. doi: 10.1001/archophthalmol.2011.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern A, Schmidt K, Leder C, Muller OJ, Wobus CE, Bettinger K, Von der Lieth CW, King JA, Kleinschmidt JA. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J Virol. 2003;77:11072–11081. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe DM, Howley PM. Fields virology. 6th edn. Philadelphia, PA; Wolters Kluwer/Lippincott Williams & Wilkins Health; 2013. [Google Scholar]
- Koerber JT, Jang JH, Schaffer DV. DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Mol Ther. 2008;16:1703–1709. doi: 10.1038/mt.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Asokan A, Wu Z, Van Dyke T, DiPrimio N, Johnson JS, Govindaswamy L, Agbandje-McKenna M, Leichtle S, Redmond DE., Jr Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Molecular therapy : the journal of the American Society of Gene Therapy. 2008;16:1252–1260. doi: 10.1038/mt.2008.100. [DOI] [PubMed] [Google Scholar]
- Lisowski L, Dane AP, Chu K, Zhang Y, Cunningham SC, Wilson EM, Nygaard S, Grompe M, Alexander IE, Kay MA. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochrie MA, Tatsuno GP, Christie B, McDonnell JW, Zhou S, Surosky R, Pierce GF, Colosi P. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J Virol. 2006;80:821–834. doi: 10.1128/JVI.80.2.821-834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis Jeune V, Joergensen JA, Hajjar RJ, Weber T. Pre-existing anti-adeno-associated virus antibodies as a challenge in AAV gene therapy. Hum Gene Ther Methods. 2013;24:59–67. doi: 10.1089/hgtb.2012.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008;320:1632–1635. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- MacLaren RE, Groppe M, Barnard AR, Cottriall CL, Tolmachova T, Seymour L, Clark KR, During MJ, Cremers FP, Black GC, et al. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet. 2014;383:1129–1137. doi: 10.1016/S0140-6736(13)62117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, Pugh EN, Jr, Mingozzi F, Bennicelli J, Banfi S, Marshall KA, Testa F, Surace EM, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshri N, Koerber JT, Kaspar BK, Schaffer DV. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nature biotechnology. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- Mays LE, Wang L, Tenney R, Bell P, Nam HJ, Lin J, Gurda B, Van Vliet K, Mikals K, Agbandje-McKenna M, et al. Mapping the structural determinants responsible for enhanced T cell activation to the immunogenic adeno-associated virus capsid from isolate rhesus 32.33. J Virol. 2013;87:9473–9485. doi: 10.1128/JVI.00596-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F, Maus MV, Hui DJ, Sabatino DE, Murphy SL, Rasko JE, Ragni MV, Manno CS, Sommer J, Jiang H, et al. CD8(+) T-cell responses to adeno-associated virus capsid in humans. Nat Med. 2007;13:419–422. doi: 10.1038/nm1549. [DOI] [PubMed] [Google Scholar]
- Nakai H, Fuess S, Storm TA, Muramatsu S, Nara Y, Kay MA. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J Virol. 2005;79:214–224. doi: 10.1128/JVI.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai H, Thomas CE, Storm TA, Fuess S, Powell S, Wright JF, Kay MA. A limited number of transducible hepatocytes restricts a wide-range linear vector dose response in recombinant adeno-associated virus-mediated liver transduction. J Virol. 2002;76:11343–11349. doi: 10.1128/JVI.76.22.11343-11349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of molecular biology. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Perabo L, Buning H, Kofler DM, Ried MU, Girod A, Wendtner CM, Enssle J, Hallek M. In vitro selection of viral vectors with modified tropism: the adeno-associated virus display. Mol Ther. 2003;8:151–157. doi: 10.1016/s1525-0016(03)00123-0. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Schnepp BC, Johnson PR. Adeno-associated virus delivery of broadly neutralizing antibodies. Curr Opin HIV AIDS. 2014;9:250–256. doi: 10.1097/COH.0000000000000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Storm T, Kay MA. Characterization of the relationship of AAV capsid domain swapping to liver transduction efficiency. Mol Ther. 2007;15:1955–1962. doi: 10.1038/sj.mt.6300293. [DOI] [PubMed] [Google Scholar]
- Vandenberghe LH, Breous E, Nam HJ, Gao G, Xiao R, Sandhu A, Johnston J, Debyser Z, Agbandje-McKenna M, Wilson JM. Naturally occurring singleton residues in AAV capsid impact vector performance and illustrate structural constraints. Gene therapy. 2009;16:1416–1428. doi: 10.1038/gt.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberghe LH, Wang L, Somanathan S, Zhi Y, Figueredo J, Calcedo R, Sanmiguel J, Desai RA, Chen CS, Johnston J, et al. Heparin binding directs activation of T cells against adenoassociated virus serotype 2 capsid. Nat Med. 2006;12:967–971. doi: 10.1038/nm1445. [DOI] [PubMed] [Google Scholar]
- Wang L, Calcedo R, Wang H, Bell P, Grant R, Vandenberghe LH, Sanmiguel J, Morizono H, Batshaw ML, Wilson JM. The pleiotropic effects of natural AAV infections on liver-directed gene transfer in macaques. Mol Ther. 2010;18:126–134. doi: 10.1038/mt.2009.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Xiao W, Conlon T, Hughes J, Agbandje-McKenna M, Ferkol T, Flotte T, Muzyczka N. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J Virol. 2000;74:8635–8647. doi: 10.1128/jvi.74.18.8635-8647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman MS. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci U S A. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular biology and evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.