Abstract
In the axial channels of ClpX and related hexameric AAA+ protein-remodeling rings, the pore-1 loops are thought to play important roles in engaging, mechanically unfolding, and translocating protein substrates. How these loops perform these functions and whether they also prevent substrate dissociation to ensure processive degradation by AAA+ proteases are open questions. Using ClpX pore-1-loop variants, single-molecule force spectroscopy, and ensemble assays, we find that the six pore-1 loops function synchronously to grip and unfold protein substrates during a power stroke but are not important in preventing substrate slipping between power strokes. The importance of grip strength is task dependent. ClpX variants with multiple mutant pore-1 loops translocate substrates as well as the wild-type enzyme against a resisting force but show unfolding defects and a higher frequency of substrate release. These problems are magnified for more mechanically stable target proteins, supporting a threshold model of substrate gripping.
Introduction
AAA+ proteolytic machines couple ATP hydrolysis to the mechanical unfolding and subsequent translocation of protein substrates into an internal chamber where degradation occurs (Sauer and Baker, 2011). The AAA+ ClpXP protease consists of an ATP-fueled unfolding and translocation motor (ClpX) and a barrel-shaped peptidase (ClpP) (Baker and Sauer, 2012). ClpX functions as a topologically closed hexameric ring with an axial channel or pore (Glynn et al., 2009; 2012). This pore is lined with loops that initially interact with the intrinsically disordered ssrA tag of a protein substrate and also play key roles in its subsequent unfolding and translocation (Martin et al., 2008a; 2008b). Notably, the GYVG or pore-1 loops (p1 loops hereafter) play roles in all stages of substrate engagement and processing by ClpX, and homologous loops are present in all AAA+ proteases and protein-remodeling machines (Siddiqui et al., 2004; Martin et al., 2008a; 2008b; Iosefson et al., 2015). ATP binding, hydrolysis, and product release are thought to drive movements of the p1 loops that translocate the degradation tag through the axial channel, ultimately forcing unfolding of the attached native protein. Additional cycles of ATP hydrolysis then power translocation of the denatured polypeptide into the ClpP chamber for proteolysis. All AAA+ proteases employ related mechanisms to unfold and degrade specific substrates to maintain protein homeostasis and quality control.
The aromatic side chain of Tyr153 in the p1 loop of Escherichia coli ClpX plays a critical functional role (Siddiqui et al., 2004). In previous work, we constructed single-chain ClpX hexamers with all possible configurations of wild-type and mutant p1 loops (Tyr153 replaced by alanine, a small apolar residue) and assayed their ability to support ClpP degradation of substrates with different stabilities (Iosefson et al., 2015). We found that neighboring p1 loops function synergistically and that the number of wild-type loops required for robust degradation increased with substrate stability. For example, a variant with three mutant loops degraded an unfolded substrate better than the parental enzyme but could not degrade a difficult to unfold substrate, native GFP-ssrA. These mutations caused degradation defects even when positioned in subunits that could not hydrolyze ATP. To explain these results, we proposed that the six p1 loops in the ClpX ring function coordinately in gripping the substrate and moving synchronously during each power stroke. The architecture of ClpX allows all six p1 loops to move in unison during a power stroke, as conformational changes initiated by ATP hydrolysis in one subunit propagate around the entire ring via coupled rigid-body interactions (Glynn et al., 2012).
Here, we characterize the properties of ClpX variants bearing p1-loop mutations in one, two, or three adjacent subunits by single-molecule force spectroscopy and find that each variant translocates substrates at rates very similar to the parental enzyme. However, these mutants have unfolding defects that are highly substrate dependent, show defects in processive degradation of multi-domain substrates, and release single-domain substrates more rapidly than the wild-type enzyme. In aggregate, our results provide additional support for a model in which the p1 loops function in a highly coordinated manner to grip substrates during the power strokes responsible for unfolding and translocation but suggest that other parts of the axial pore, possibly the pore-2 loops, play a major role in gripping the substrate between the completion of one power stroke and the beginning of the next power stroke.
Results
Optical-trap assays of unfolding
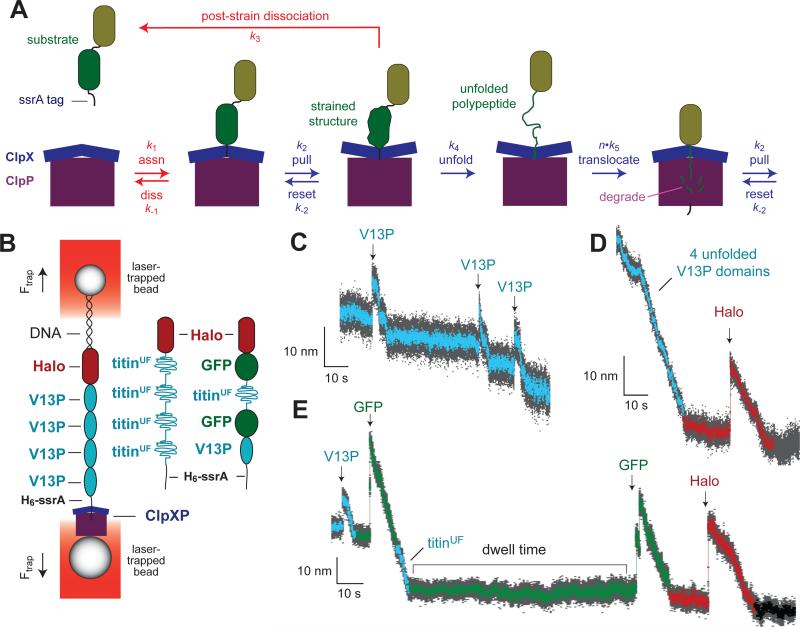
Fig. 1A shows a minimal model for ClpXP degradation of a multi-domain substrate. Steps colored blue determine the rates of single-molecule unfolding and translocation, whereas these steps in addition to the steps colored red contribute to ensemble degradation in solution. For single-molecule studies, we used covalently linked ClpXΔN hexamers with a single biotinylation site and Y153A p1-loop mutations in one (AYYYYY), two (AAYYYY), or three (AAAYYY) subunits, as well as the parental YYYYYY enzyme (Iosefson et al., 2015). Biotinylated ClpX or ClpXP was attached to one streptavidin-coated bead and a protein substrate with a C-terminal ssrA tag and an N-terminal Halo-domain was attached to a second bead via a double-stranded DNA linker (Fig. 1B; Aubin-Tam et al., 2011). Substrates included Halo-(V13P)4-H6-ssrA, Halo-(V13PCM)4-H6-ssrA, and Halo-GFP-titinUF-GFP-V13P-H6-ssrA, where V13P signifies a native titinI27 domain containing the destabilizing Val13→Pro mutation, V13PCM is the same domain permanently unfolded by carboxymethylation of cysteines in the hydrophobic core, titinUF is a titinI27 domain denatured by Cys→Asp core mutations, and GFP is green fluorescent protein. Representative single-molecule trajectories of unfolding and translocation of these substrates by ClpXP p1-loop variants are shown in Figs. 1C, 1D, and 1E. In these traces, sudden increases in bead-to-bead distance mark unfolding events, decreases in bead-to-bead distance represent translocation, and periods of steady bead-to-bead distance correspond to a pre-unfolding dwell in which denaturation attempts are unsuccessful.
Figure 1.
ClpXP degradation. (A) Model for ClpXP degradation of a two-domain protein bearing an ssrA degradation tag. Blue arrows are kinetic steps that contribute to unfolding and translocation in solution degradation and in optical-trap experiments, whereas red arrows are steps that only contribute to degradation in ensemble assays. The k1 association step requires ATP binding but not hydrolysis. Mechanical steps (k2 and k5) are power strokes fueled by ATP hydrolysis. In an optical-trap experiment, dissociation by the k−1 or k3 pathways would terminate the experiment. Similarly, following degradation of the green domain, release of the olive domain by a k3-like step would end an optical-trap experiment. (B) Cartoon of optical-trap experiment. ClpXP bound to one laser-trapped bead engages a multi-domain substrate bound to a second laser-trapped bead, creating a stretched tether that offsets a force that would otherwise move the beads apart as a consequence of the laser positions and power. The beads move farther apart when ClpXP unfolds a native domain and closer together when ClpXP translocates an unfolded segment of the substrate. These changes in bead-to-bead distance provide an assay of the mechanical activities of ClpXP. Different substrates used in optical-trap studies are depicted schematically. (C) Optical-trap trace of AAAYYY ClpXP degradation of the Halo-(V13P)4-H6-ssrA substrate, showing three V13P unfolding events (arrows) and subsequent translocation at an experimental force of 5.2 pN. In this and subsequent panels, bead-to-bead distances decimated to 300 Hz are gray and those decimated to 30 Hz are colored. (D) Trace of AAYYYY ClpXP degradation of the Halo-(V13PCM)4-H6-ssrA substrate at 7 pN. Cyan symbols correspond to translocation of the unfolded V13P titin domains, whereas dark-red symbols show the pre-unfolding dwell time, unfolding (arrow), and translocation of the Halo domain. (E) Trace of AYYYYY ClpXP degradation of the Halo-GFP-titinUF-GFP-V13P-ssrA substrate (8 pN). Cyan symbols represent titin events, green symbols represent GFP events, and dark-red symbols represent Halo events.
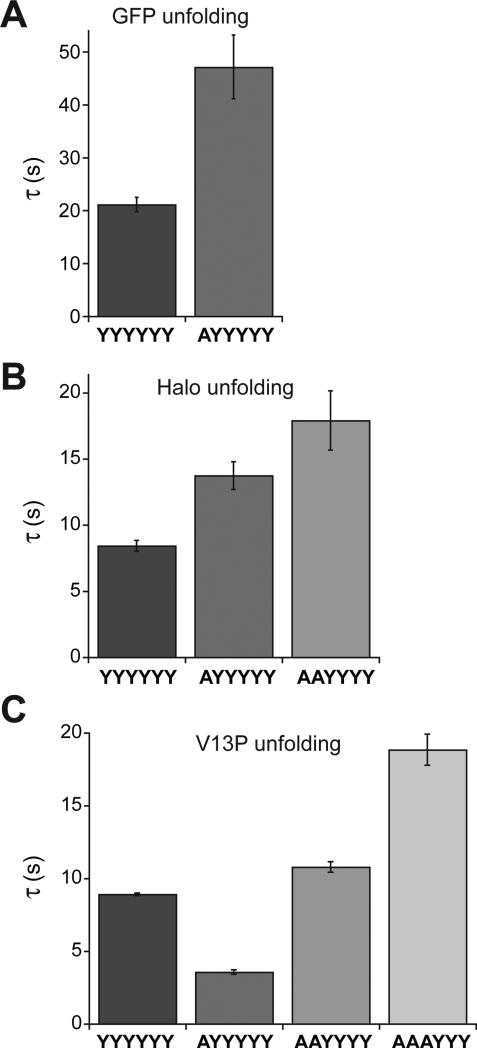
Previous studies using the parental ClpXP enzyme revealed that the pre-unfolding dwell times for a given domain are exponentially distributed with a time constant that reflects mechanical stability (Aubin-Tam et al., 2011; Maillard et al., 2011; Sen et al., 2013; Cordova et al., 2014; Olivares et al., 2014). The pre-unfolding dwell times for the V13P, Halo and GFP domains in our substrates were also distributed exponentially (Fig. S1), as expected if each power stroke has a constant probability of successful unfolding. In general, the average pre-unfolding dwell for a specific domain increased as the number of wild-type p1 loops in the ClpX variant decreased (Fig. 2; Fig. S1), especially for domains more resistant to unfolding. For unfolding of GFP (Fig. 2A), for example, the average unfolding dwells (τ) were ~21 s for YYYYYY, ~47 s for AYYYYY, and no unfolding events were recorded for AAYYYY or AAAYYY. For unfolding of the Halo domain (Fig. 2B), the τ values were approximately 8 s (YYYYYY), 14 s (AYYYYY), and 18 s (AAYYYY), with too few recorded unfolding events by AAAYYY (N = 3) to allow fitting. For unfolding of the V13P titin domain (Fig. 2C), the τ values were roughly 9 s (YYYYYY), 4 s (AYYYYY), 11 s (AAYYYY), and 18 s (AAAYYY). Thus, for this domain, the trend of increasing τ values with increasing p1-loop mutations is reversed for the YYYYYY and AYYYYY enzymes (see Discussion).
Figure 2.
Average pre-unfolding dwell times (τ) for GFP domains (A), Halo domains (B), and V13P-titin domains (C) were determined from single-exponential fits of the dwell-time distributions shown in Fig. S1. Errors are ± SEM of the fits.
Single-molecule translocation
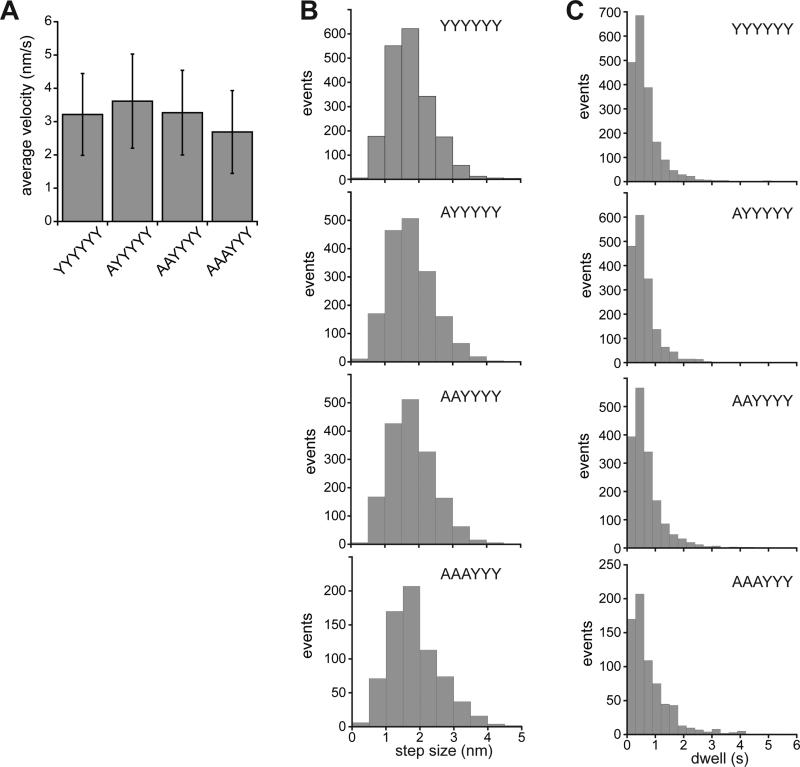
For each enzyme, we calculated average translocation velocities for the titin V13P domains in the Halo-(V13P)4-H6-ssrA and Halo-(V13PCM)4-H6-ssrA substrates without attempting to define pauses or remove them from the calculated rates (Fig. 3A). These values ± SD were 3.2 ± 1.2 nm/s (YYYYYY; N=181); 3.6 ± 1.4 nm/s (AYYYYY; N=144), 3.3 ± 1.3 nm/s (AAYYYY; N=140), and 2.7 ± 1.3 nm/s (AAAYYY; N=60). Thus, the p1-loop mutations do not cause major changes in translocation velocity. For each enzyme, linear fits of translocation velocity against experimental force had slopes close to 0 and correlation coefficients between 0 and 0.11 (Fig. S2), suggesting weak force dependence.
Figure 3.
Translocation parameters for unfolded titin domains. (A) Average translocation velocity ± SD for YYYYYY, AYYYYY, AAYYYY, and AAAYYY ClpXP calculated from the data shown in Fig. S2. (B) Histograms showing distributions of translocation step sizes. (C) Histograms showing distributions of pre-step dwell times.
We used a step-finding algorithm (Kerssemakers et al., 2006) to determine step sizes and pre-step dwell times during translocation of titin domains by the variants and the parental enzyme. As previously reported (Aubin-Tam et al., 2011; Maillard et al., 2011; Sen et al., 2013; Cordova et al., 2014), the YYYYYY parent took translocation steps ranging from ~1 to ~4 nm in length. Notably, the p1-loop variants displayed step-size distributions (Fig. 3B) and distributions of pre-step dwell times (Fig. 3C) very similar to the parent, with no evidence that the variants pause significantly more frequently. For each enzyme, a lag in the distribution of pre-step dwell times suggested that multiple kinetic steps contribute to most physical steps, consistent with longer steps resulting from a burst of individual ~1-nm power strokes (Sen et al., 2013; Cordova et al., 2104). An average translocation velocity of 3 ± 1 nm/s in the optical trap at room temperature corresponds to an average ATP-hydrolysis rate of 180 ± 60 min−1, assuming that 1-nm steps result from hydrolysis of one ATP, 2-nm steps result from hydrolysis of two ATPs, etc. Thus, the similar translocation rates of the parent and variants indicate that they hydrolyze ATP at similar rates. Indeed, prior studies show ATP-hydrolysis rates of 235 ± 35 min−1 during degradation of an unfolded substrate by these enzymes in ensemble studies at 30 °C (Iosefson et al., 2015).
Single-molecule detection of substrate slipping
If a substrate slips from the grasp of ClpXP in the optical trap, the bead-to-bead distance increases just as it does upon domain unfolding. A key difference, however, is that the length of a slip is not limited, whereas the length of an unfolding increase is constrained by the number of amino acids in the domain. Because of degradation, the enzyme-engaged tail of the protein substrate is ~35 residues (Lee et al., 2001; Kenniston et al., 2005; Martin et al., 2008b), and slipping of more than this number of residues breaks the bead-to-bead tether and terminates the experiment. To ask if tethers break more frequently for the p1-loop variants than the parent, we calculated the average duration ± SEM of single-molecule traces for the Halo-(V13P)4-H6-ssrA substrate. These durations were 65 ± 6 s (YYYYYY), 47 ± 6 s (AYYYYYY), 54 ± 8 s (AAYYYY), and 101 ± 24 s (AAAYYY). An average of 2.5-2.9 V13P-unfolding-events/trace was also recorded for the parent and mutants. Thus, single-molecule tethers supported by the p1-loop variants do not appear to be dramatically more fragile than those supported by parental ClpXP, although multiple factors, including different unfolding activities, contribute to these average trace durations.
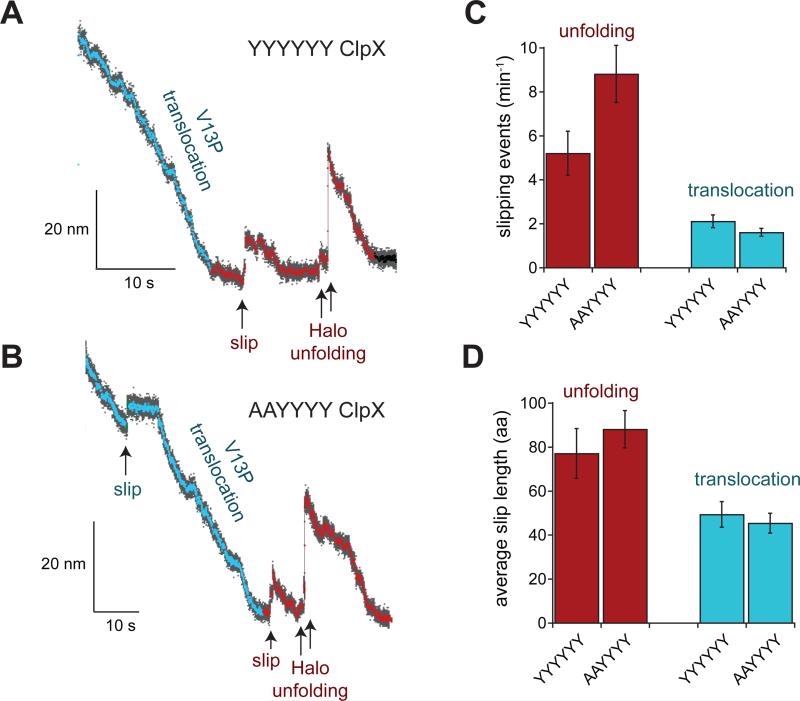
Slipping in the optical trap is more readily assayed using ClpX in the absence of ClpP, as most slips along an undegraded substrate do not break the bead-to-bead tether (Aubin-Tam et al., 2011; Maillard et al., 2011). Thus, we performed trap assays using the Halo-(V13PCM)4-H6-ssrA substrate and the YYYYYY and AAYYYY ClpX variants without ClpP. The distributions of translocation step sizes and translocation pre-step dwells were similar for both enzymes (Fig. S3) and similar to the ClpXP distributions (cf. Figs. S3 & 3). Individual traces with slipping events during translocation or the Halo pre-unfolding dwell period are shown for YYYYYY ClpX in Fig. 4A and for AAYYYY ClpX in Fig. 4B. At forces averaging ~10 pN (range 6-14 pN), the AAYYYY variant slipped ~50% more frequently during attempts to unfold the Halo domain than did the parental YYYYYY enzyme (Fig. 4C), but the average slip length (~80 amino acids) was similar (Fig. 4D). Thus, the effect of removing two wild-type p1 loops on substrate slipping is not dramatic. During translocation of the titin domain, the average frequencies and slip lengths were similar for both enzymes, although both parameters were reduced compared to unfolding slips (Figs. 4C, 4D). Thus, ClpX slips more frequently during unfolding attempts than during translocation, and a full set of wild-type p1 loops appears to reduce slipping during unfolding.
Figure 4.
Slipping during translocation and unfolding of the Halo-(V13PCM)4-H6-ssrA substrate by the YYYYYY and AAYYYY ClpX enzymes in the absence of ClpP. Data were recorded at average forces of ~10 pN (range 6-14 pN) for both YYYYYY and AAYYYY. (A) Individual trace with a slipping event for YYYYYY ClpX during the pre-unfolding dwell in an optical-trap experiment at 6.5 pN. Bead-to-bead distances decimated to 300 Hz are shown in gray and to 30 Hz in color. In this trace, the Halo domain unfolds in two steps, probably corresponding to extraction of the C-terminal helix followed by unfolding of the remaining structure. Similar two-step Halo unfolding was observed in 70-80% of the traces with ClpX alone or with ClpXP. (B) Individual trace with slipping events during translocation and the pre-unfolding dwell by AAYYYY ClpX in an experiment at 12.5 pN. Decimation and coloring as in panel A. (C) Number of slipping events min−1 for YYYYYY and AAYYYY ClpX during the Halo pre-unfolding dwell and translocation. (D) Average slip length for YYYYYY and AAYYYY ClpX during the Halo pre-unfolding dwell and translocation. In panels C and D, values are the mean ± SD of averages calculated for 35 independent trials in which 33% of the data was randomly discarded.
A longer unstructured tail compensates for reduced grip
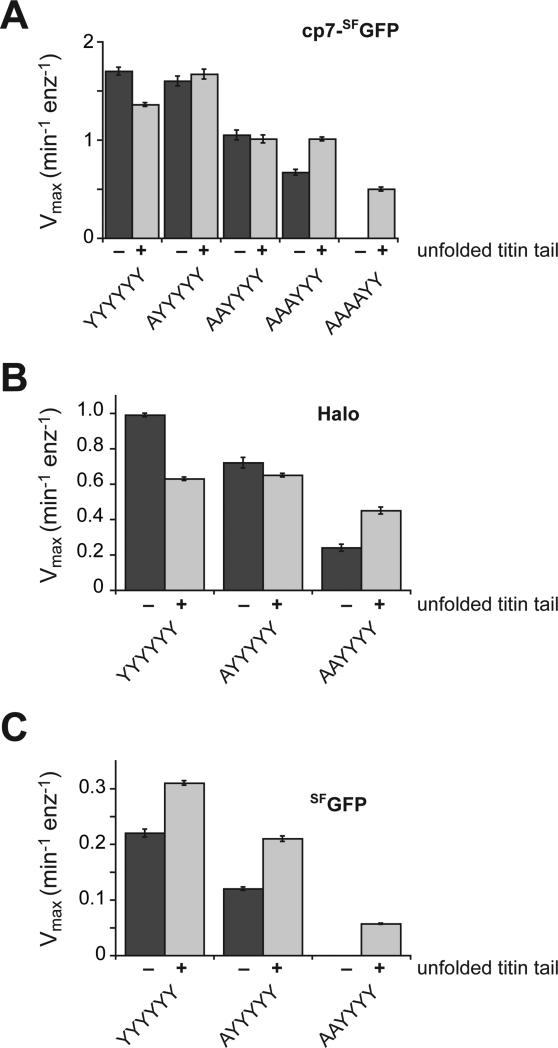
ClpXP degrades GFP-V13PCM-ssrA faster than GFP-ssrA, despite the extra time required to translocate ~100 additional amino acids of the unstructured V13PCM segment (Martin et al., 2008c). When ClpX encounters the native domain in GFP-V13PCM-ssrA, faster unfolding likely results from improved grip on a longer segment of polypeptide in the axial pore.1 We performed similar experiments to ask if more polypeptide in the ClpX pore might compensate for reduced grip caused by pore-loop mutations. For a relatively easily unfolded substrate, cp7-SFGFP (Nager et al., 2011), an adjacent V13PCM sequence slowed degradation by YYYYYY ClpXP (probably because of increased substrate length), had little effect on degradation by AYYYYY and AAYYYY ClpXP, and accelerated degradation by AAAYYY ClpXP (Fig. 5A; Table S1; Fig. S4). Notably, AAAAYY ClpXP degraded cp7-SFGFP-V13PCM-ssrA reasonably well but could not degrade cp7-SFGFP-ssrA. For Halo, a domain of moderate mechanical stability (Popa et al., 2013), an adjacent V13PCM sequence slowed degradation by YYYYYY ClpXP, had little effect on degradation by AYYYYY ClpXP, and increased the rate of degradation by AAYYYY ClpXP (Fig. 5B; Table S1). For SFGFP, a domain very difficult to unfold (Nager et al., 2011), an adjacent V13PCM sequence increased the rate of degradation by YYYYYY and AYYYYY ClpXP, and allowed degradation by AAYYYY ClpXP, which could not degrade SFGFP-ssrA (Fig. 5C; Table S1). Thus, a longer region of unfolded polypeptide in the ClpX pore can compensate for p1-loop unfolding defects, although improved unfolding may depend upon the sequence and length of the polypeptide in the pore (Too et al., 2013).
Figure 5.
Tail-length effects on solution degradation. Plots of Vmax for steady-state degradation of a cp7-SFGFP domain (A), a Halo domain (B), or a SFGFP domain (C) in substrates with a C-terminal ssrA tag (−) or an unfolded C-terminal V13PCM-ssrA tag (+) by parental ClpXP and different p1-loop variants. Values are averages (N = 3) ± SEM. Initial rates of degradation of different concentrations of substrate by ClpX variants (0.1 μM), ClpP (0.3 μM), and ATP (4 mM) were determined and fit to the Michaelis-Menten equation to determine KM and Vmax values (Fig. S4) or were taken from Iosefson et al. (2015) (Table S1).
P1-loop variants release stable protein domains more frequently
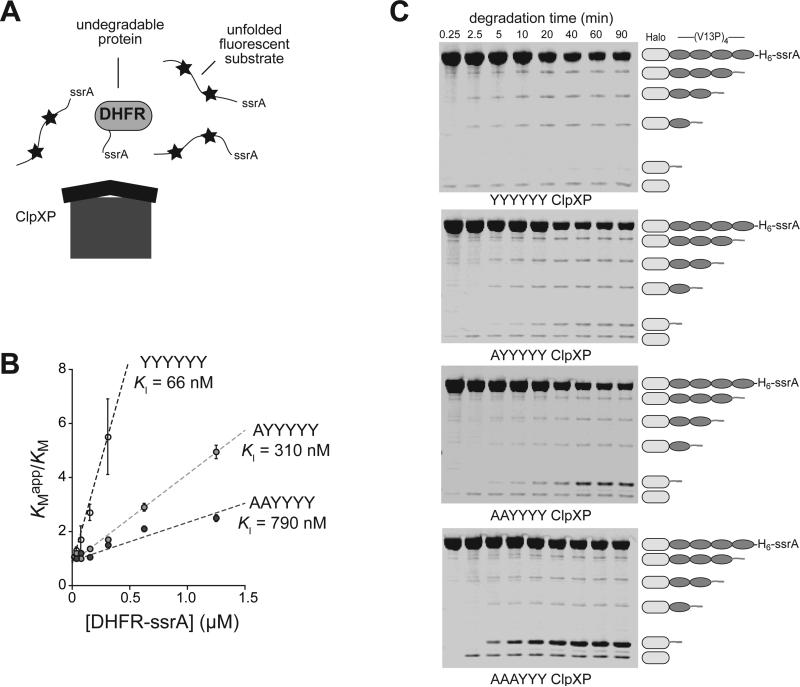
Stable substrates can be bound and released multiple times from ClpXP before unfolding is successful and degradation begins (Kenniston et al., 2005). To test if p1-loop variants release stable substrates more frequently than the parent enzyme, we inhibited degradation of fluorescent unfolded titin-ssrA using methotrexate-bound E. coli DHFR-ssrA, which cannot be unfolded by ClpXP (Fig. 6A; Lee et al., 2001; Too et al., 2013). As expected, increasing MTXDHFR-ssrA resulted in competitive inhibition of degradation of unfolded titin by YYYYYY, AYYYYY and AAYYYY ClpXP, increasing KMapp without affecting Vmax (Fig. S5). To determine constants (KI) for DHFR-ssrA inhibition of each enzyme, KMapp for each inhibitor concentration was divided by KM in the absence of competitor and plotted against the inhibitor concentration (Fig. 6B). Because KMapp/KM = 1 + [MTXDHFR-ssrA]/KI, the inhibition constant is the reciprocal of the slope of a linear fit. Importantly, MTXDHFR-ssrA was a substantially better inhibitor for YYYYYY ClpXP (KI = 66 nM) than for AYYYYY ClpXP (KI = 310 nM) or AAYYYY ClpXP (KI = 790 nM). These results strongly support a model in which non-degradable DHFR-ssrA is a better inhibitor of parental ClpXP because it is released more readily by the p1-loop variants.
Figure 6.
Effects of p1-loop mutations on release of undegraded protein domains. (A) In the presence of methotrexate, DHFR-ssrA cannot be degraded by ClpXP and acts as a competitive inhibitor of the degradation of an unfolded ssrA-tagged substrate labeled with fluorescent dyes (stars), which are auto-quenched in the undegraded protein. (B) KMapp/KM for degradation of the fluorescent unfolded substrate by YYYYYY, AYYYYY, and AAYYYY ClpXP plotted against the concentration of the DHFR-ssrA competitor. Values are averages (N = 3) ± SEM. The lines are linear fits to KMapp/KM = 1 + [DHFR-ssrA]/KI. Degradation rates and apparent KM and Vmax values for different concentrations of the fluorescent substrate by ClpX variants (0.1 μM), ClpP (0.3 μM), and ATP (4 mM) were determined in the absence of competitor and in the presence of different concentrations of competitor (Fig. S3). (C) Degradation of TAMRA-Halo-(V13P)4-ssrA by the parental ClpXP and the p1-loop variants was assayed by SDS-PAGE. Fluorescent bands corresponding to TAMRA-containing fragments were detected using a Typhoon imager. Degradation reactions were performed at 30 °C and contained ClpX variants (1 μM hexamer), ClpP (2 μM tetradecamer), fluorescent substrate (10 μM), and ATP (10 mM). The lowest band on the gel was observed in a control experiment without ATP, suggesting that it arises from contaminating protease activity. Although larger fragments should appear faster in these experiments, those containing three, two, or one V13P domains were found to arise at similar rates. This behavior is expected for a reaction in which the rate of substrate engagement is slow compared to the rate at which individual V13P domains are degraded, especially if substrate release upon encounter with a V13P domain is rare.
To assess the effects of p1-loop grip on substrate release in the midst of degradation, we labeled the Halo domain of Halo-(V13P)4-H6-ssrA with a fluorescent TAMRA dye and monitored the kinetics of degradation by SDS-PAGE followed by detection of fluorescent proteins (Fig. 6C; Fig. S6). In this assay, substrate release after degradation has begun from the C-terminal ssrA tag results in the accumulation of partially degraded fluorescent species lacking an ssrA tag. For YYYYYY ClpXP, all partially degraded species accounted for only ~1% of the protein degraded by 90 min, indicating that parental ClpXP degradation proceeds through the V13P and Halo domains in a highly processive fashion. During degradation by AYYYYY, AAYYYY, and AAAYYY ClpXP, partially degraded species containing V13P domains accounted for 2-3% of the protein degraded at 90 min, indicating an increased level of release but still highly processive degradation. Notably, after 90 min, a product corresponding to the Halo domain with a ~35-residue tail accounted for approximately 1%, 6%, and 60% of the degraded substrate for AYYYYY, AAYYYY and AAAYYY ClpXP, respectively. Thus, a full complement of p1 loops is not necessary for highly processive degradation of V13P domains, but release of the more mechanically stable Halo domain becomes more probable in the midst of degradation as the number of mutant pore loops increases.
Discussion
The p1 loop of ClpX is a GYVG sequence that connects two α-helices, which are part of a rigid-body unit (Kim & Kim, 2003; Glynn et al., 2009). Thus, these loops in the ClpX ring move as part of rigid-body motions initiated by ATP binding, hydrolysis, or product release in different subunits (Glynn et al., 2012). As the number of mutant p1 loops in a ClpX hexamer increases, we previously found that ClpXP progressively loses the ability to degrade native substrates with increased mechanical stability, probably because of defects in initiating unfolding (Iosefson et al., 2015). As shown in Fig. 1A, however, degradation requires multiple kinetic steps, and it is difficult to confidently ascribe changes in rates of ensemble proteolysis to a specific reaction step. In optical-trap experiments, by contrast, translocation and unfolding can be directly visualized. For example, AAAYYY ClpXP degrades unfolded titin in ensemble experiments about twice as fast and at half the ATP cost as parental ClpXP, but whether AAAYYY translocates the substrate faster or engages the substrate more efficiently was uncertain (Iosefson et al., 2015). Because translocation in the optical trap is virtually identical for AAAYYY and YYYYYY ClpXP, however, it now appears that the mutant engages the substrate more efficiently. Several lines of evidence suggest that the axial pore is elastic, expanding to accommodate larger polypeptides and contracting to maintain grip on skeletal substrates (Barkow et al., 2009; Glynn et al., 2009). The axial pore in substrate-free ClpX would be closed, by this model, and would have to open to allow substrate engagement, which could be easier to accomplish for the less tightly packed pore of the AAAYYY variant.
A subset of functional p1 loops supports normal translocation
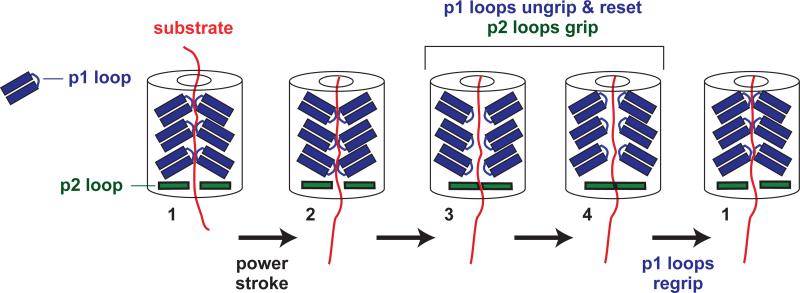
In one model of ClpX function, some p1 loops in the ring move during a power stroke, while others remain static to grip the polypeptide and prevent slipping (for example, see Fig. 3 in Martin et al., 2008b). This model predicts that p1-loop variants should translocate more slowly, take shorter steps, or pause or slip more frequently during translocation. By contrast, we find that ClpX variants with one, two, or three p1-loop mutations translocate polypeptides with essentially the same distribution of translocation-step sizes and pre-step dwell times as the parent, yielding similar average translocation velocities. The frequency and length of slipping events during translocation is also similar for YYYYYY and AAYYYY ClpX. In combination with previous studies, these results support a model in which the six p1 loops of ClpX function coordinately to translocate or unfold the substrate (Iosefson et al., 2015). In the model of Fig. 7, for example, all six p1 loops contact the substrate and move in concert during a power stroke that drives a 1-nm translocation step. A 2-nm step requires a kinetic burst of two power strokes, etc. Concerted p1-loop movement during a single power stroke does not require concerted or sequential ATP hydrolysis in the ClpX ring, and multiple experiments support probabilistic ATP hydrolysis by individual subunits (Martin et al., 2005; Stinson et al., 2015). Therefore, each stochastic hydrolysis event appears to result in a 1-nm power stroke in which all of the p1-loops move.
Figure 7.
Cartoon model for substrate gripping by ClpX. During a power stroke (conformation 1 → 2), substrate grip is maintained by the six p1 loops of the hexamer, which move in concert to translocate the polypeptide. The p1 loops are shown acting as opposed pairs but could be arranged helically or in other geometries. After the initial power stroke (conformations 3 and 4), the p1 loops release the polypeptide and grip is maintained by the p2 loops (only two of six shown for simplicity). In the conformation 4 → 1 transition, the p1 loops regrip and the p2 loops release the substrate in preparation for the next power stroke. The cycle shown results from hydrolysis of one ATP and translocates the polypeptide ~1 nm (~5 amino acids). Longer translocation steps result from kinetic bursts of multiple power strokes. A similar model could account for maintaining grip during substrate unfolding.
In the optical trap, ClpXP translocates against a resisting force of ~5-20 pN, and yet very little slipping of the substrate is observed for the parent or for the p1-loop mutants. If all six p1 loops move during a power stroke, then how is slipping prevented when these loops have to release and regrip the substrate before the next power stroke? One possibility is that other parts of the axial pore of ClpX, possibly supported by ClpP, maintain substrate grip during the resetting reaction (Fig. 7). The pore-2 loops are a candidate for this role, as they crosslink to substrate, participate in unfolding, and interact with ClpP (Martin et al., 2007; Martin et al., 2008b).
P1-loop function during unfolding
Although as many as three p1 loops in the ClpX hexamer can be mutated without compromising translocation, substantial differences in single-molecule unfolding of different protein domains by parental ClpXP and the p1-loop variants are seen. For example, a mutation in the p1 loop of a single ClpX subunit roughly doubled the time required to unfold GFP. For the Halo domain, we also observed progressively slower single-molecule unfolding for ClpXP variants with an increased number of mutant p1 loops. The trend of decreasing unfolding activity with increasing numbers of p1-loop mutations did not hold for the V13P domain, which YYYYYY ClpXP unfolded more slowly than AYYYYY and only ~20% faster than AAYYYY. As V13P is the least mechanically stable domain investigated, unfolding grip strength may only become important above a threshold, analogous to the grip between a vehicle tire and a wet road becoming more important at increased speeds. In ensemble degradation, AYYYYY ClpXP also degraded cp7-SFGFP-V13PCM-ssrA slightly faster than the parent enzyme. Thus, removing one wild-type pore loop in AYYYYY may enhance unfolding of unstable domains by a mechanism that does not involve grip. For example, the AYYYYY enzyme might access a different unfolding pathway for V13P, as the data were fit best by a double exponential with one time constant faster and one slower than the wild-type value (Fig. S1). Better unfolding by a similar mechanism in the AAYYYY and AAAYYY variants could be offset by the more severe grip defects in these variants. The important result, however, is that variants with more p1-loop mutations usually unfold a given domain more slowly.
To unfold mechanically stable substrates, enzymes that grip the protein less tightly require a greater average number of power strokes. In the model of Fig. 1A, the rate constants for pulling via a power stroke (k2), resetting the ring for the next power stroke (k−2), and unfolding (k4) determine the average time and number of power strokes required to unfold a domain in the optical trap. The pulling and resetting steps are correlated with the rate of ATP hydrolysis during unfolding, which cannot be measured directly in trap experiments. However, p1-loop variants translocate at near wild-type rates, suggesting that they are not defective in ATP hydrolysis, and YYYYYY, AYYYYY, AAYYYY, and AAAYYY ClpXP hydrolyze ATP at similar rates during solution degradation (Iosefson et al., 2015). Thus, p1-loop mutations appear to decrease k4, as expected if reduced grip on the substrate during a power stroke decreases the probability of unfolding.
ClpX slips more frequently during unfolding than translocation in the optical trap, probably because the force of the strained native substrate pulling back on the enzyme is added to the trap force. Additional force during unfolding may alter the structure of the ClpX ring and thus reduce substrate grip. In optical-trap experiments in the absence of ClpP, AAYYYY ClpX slipped more frequently than YYYYYY ClpX during Halo unfolding, but both enzymes slipped at a similar rate during translocation, supporting the idea that grip defects are magnified during unfolding.
Solution degradation
In ensemble degradation, intact substrates can be released from ClpXP before or after a power stroke (the k−1 and k3 steps in Fig. 1A). These dissociation steps typically slow but do not prevent degradation, as the intact substrate rebinds and is eventually unfolded and degraded. Interestingly, some degradation defects of p1-loop variants can be suppressed by changing the length or sequence of the substrate in the axial pore during unfolding. For example, inserting an unfolded V13P domain between the ssrA tag and a difficult to unfold protein domain allows AAYYYY and AAAAYY ClpXP to degrade substrates that they otherwise could not degrade. In these cases, the unfolded V13P sequence in the ClpX pore compensates for reduced grip caused by the p1-loop mutations. Notably, however, improved grip only becomes important as the mechanical stability of the substrate increases and/or the number of p1-loop mutations increases, again supporting a threshold effect.
How well a protein substrate remains bound to ClpXP when unfolding is unsuccessful also appears to be a function of p1-loop grip strength. For example, MTXDHFR-ssrA, which cannot be unfolded by ClpXP, competitively inhibits degradation of another protein far more efficiently for parental ClpXP than for p1-loop variants, as expected if the mutants release MTXDHFR-ssrA more frequently after a power stroke.
Release of a target protein after proteolysis of its degron prevents further ClpXP degradation, providing an assay for partitioning between release and unfolding/degradation (Lee et al., 2001; Kenniston et al., 2005; Koodathingal et al., 2009). For the native V13P titinI27 domain, we find that parental ClpXP and the AYYYYY, AAYYYY, and AAAYYY variants almost never release the substrate, resulting in highly processive degradation. For the Halo domain, by contrast, the native domain is released by ClpXP with increasing frequency as the number of mutant p1-loops increases. This result further supports a model in which partitioning between substrate unfolding and release is a function both of the mechanical stability of the native domain and of the grip strength of ClpXP and its variants.
For ssrA-tagged proteins, release during initial engagement and attempted unfolding by ClpXP is more frequent than during translocation, ensuring that degradation of single-domain substrates is highly processive once unfolding occurs. Release of difficult to unfold substrates would also allow ClpXP to engage and degrade less-stable substrates in the cell, prioritizing proteolysis of more easily degraded proteins (Kenniston et al., 2005). After ClpXP degrades the first domain of a multi-domain substrate, there is some probability of release that depends on the mechanical stability of each native domain it then encounters (Lee et al., 2001; Koodathingal et al., 2009; Too et al., 2013). This mechanism allows ClpXP to partially degrade and then release a natural substrate in a truncated form with a new biological activity (Vass and Chien, 2013). The 26S proteasome also truncates specific substrates, almost certainly by similar mechanisms (Tian et al., 2005; Kraut et al., 2012). Because ClpXP must degrade thousands of different proteins in the cell, the ability of the pore-1 loops of ClpX to grip substrates is likely to be an evolutionary compromise between efficient degradation, efficient release of intact proteins that cannot be readily unfolded, and the ability to release partially degraded protein fragments when advantageous. Similar principles are likely to apply broadly, as the pore-1 loops are highly conserved and required for the function of other AAA+ proteases, including ClpAP, HslUV, Lon, FtsH, PAN, and the Rpt1-6 unfolding ring of the 26S proteasome.
Methods
Substrates and enzymes
Construction of expression vectors for H6-tagged E. coli ClpXΔN covalent hexamers containing the Y153A p1-loop mutation, E. coli ClpP-H6, human H6-titinI27-ssrA, Halo-(V13P)4-H6-ssrA, Halo-GFP-titinUF-GFP-V13P-H6-ssrA, H6-SFGFP-ssrA, and H6-cp7-SFGFP-ssrA have been described (Nager et al., 2011; Cordova et al., 2014; Olivares et al., 2014; Iosefson et al., 2015). The nucleotide sequence encoding the full-length engineered Rhodococcus rhodochrous haloalkane dehalogenase (HaloTag) domain was amplified by PCR from plasmid pFN18A (Promega) and cloned into a pET22b (Novagen) vector (Los and Wood, 2007), with the final encoded protein containing a C-terminal H6-ssrA tag. The Halo-V13P-H6-ssrA, H6-SFGFP-V13P-ssrA, and H6-cp7-SFGFP-V13P-ssrA substrates were constructed by inserting the nucleotide sequence encoding V13P with no linker directly between the sequences encoding the Halo or GFP domain and the C-terminal ssrA or H6-ssrA tags of the corresponding substrates without the V13P sequence. A plasmid encoding E. coli DHFR was a gift from A. Matouschek (U. Texas, Austin). Using standard PCR techniques, we transferred the DHFR gene into pET22b to contain a C-terminal H6-ssrA tag.
Protein purification and labeling
Unless noted, all purification steps were performed at 4 °C. ClpX variants and ClpP were purified as reported (Iosefson et al., 2015). Fluorescently-labeled titinUF was purified and prepared as described (Iosefson et al., 2015). For purification of other proteins used in this study, cell pellets were resuspended with stirring in lysis buffer (25 mM HEPES, pH 7.6, 500 mM NaCl, 20 mM imidazole, 10% glycerol (v/v)) supplemented with benzonase and 1 mg/ml lysozyme for 1 h. One tablet of protease inhibitor mixture (Roche Applied Science) for each 100 ml of resuspended cells was also added (this step was omitted for the ClpP purification). Resuspended cells were sonicated, and the lysate was centrifuged for 1 h at 14,000 RPM. The supernatant was mixed for 30 min with Ni++-agarose beads (Qiagen) equilibrated in lysis buffer, beads were washed three times with lysis buffer, and then transferred to an empty column (Bio-Rad). The Halo-(V13P)4-H6-ssrA, Halo-GFP-titinUF-GFP-V13P-H6-ssrA, H6-SFGFP-ssrA, H6-cp7-SFGFP-ssrA, H6-SFGFP-V13P-ssrA, H6-cp7-SFGFP-V13P-ssrA, and DHFR-H6-ssrA proteins were eluted with lysis buffer supplemented with 300 mM imidazole, dialyzed overnight into 25 mM HEPES (pH 7.6), 50 mM NaCl, 0.5 mM EDTA, 10% glycerol (v/v) and further purified by ion-exchange chromatography on a MonoQ 10/100 GL column (GE Healthcare). As a final purification step, all proteins were chromatographed on a HiLoad 16/60 Superdex 200 size-exclusion column (GE Healthcare) pre-equilibrated in GF buffer (25 mM HEPES, pH 7.6, 150 mM NaCl, 10% glycerol (v/v)). DHFR-H6-ssrA was chromatographed on a HiLoad 16/60 Superdex 75 size-exclusion column (GE Healthcare) equilibrated in GF buffer. For purification of Halo-H6-ssrA and Halo-V13P-H6-ssrA, proteins were bound to Ni++-agarose beads, washed three times with lysis buffer, washed with PBS (pH 7.4), and eluted with PBS plus 300 mM imidazole (pH 7.4). HaloTag TAMRA ligand (Promega) was added to a five-fold molar excess of Halo-H6-ssrA or Halo-V13P-H6-ssrA, and mixtures were incubated for ~12 h in the dark on a rotating platform. As a final purification step, the fluorescent Halo-H6-ssrA or Halo-V13P-H6-ssrA proteins were chromatographed on a HiLoad 16/60 Superdex 200 size-exclusion column (GE Healthcare) in GF buffer. Denaturation of the V13P titin domain by carboxymethylation of its cysteines in Halo-(V13P)4-H6-ssrA, H6-SFGFP-V13P-ssrA, H6-cp7-SFGFP-V13P-ssrA, and Halo-V13P-H6-ssrA was performed following the size-exclusion purification step by reacting proteins with an excess of iodoacetic acid (Sigma) in GF buffer supplemented with 50 mM Tris-HCl (pH 8.8) for 2 h in the dark at 30 °C. Following carboxymethylation, proteins were repurified on a HiLoad 16/60 Superdex 200 size-exclusion column. Denaturation of the V13P domain was verified by observing a shift relative to the untreated native protein either in the elution volume in the size-exclusion step or in the tryptophan emission spectra. All proteins were stored frozen at −80 °C.
Single-molecule measurements
Single-molecule trajectories of substrate unfolding and translocation by ClpXP or ClpX variants were performed under constant force (18-22 °C; 4 mM ATP with ATP-regeneration and oxygen-scavenging systems) and were recorded and analyzed as described (Aubin-Tam et al., 2011; Cordova et al., 2014). The size of slips during translocation or prior to Halo unfolding was calculated using a custom MATLAB script. Translocation slips were identified as sudden increases in bead-to-bead distance ≥ 2 nm, a value greater than the 1.2 nm noise (SD) of decimated/filtered data, during translocation of the V13PCM portion of Halo-(V13PCM)4-H6-ssrA or during translocation following a slip in the Halo pre-unfolding dwell time. Slips during the period of steady bead-to-bead distance prior to Halo unfolding were identified by the same size criterion. Authentic Halo unfolding events were identified by size and/or characteristic two-step unfolding and thus distinguished from slipping events. Successive events in which ClpX slips, re-grips without translocation, and slips again were counted as a single slip. To calculate slipping frequency, we quantified the number of translocation slips or pre-unfolding slips in each trace and divided these numbers by the sum of the total translocation times or total pre-unfolding times, respectively, in that trace.
Biochemical assays
Degradation assays were performed at 30 °C in PD buffer (25 mM HEPES, pH 7.6, 100 mM KCl, 20 mM MgCl2, 10% glycerol (v/v)) supplemented with 4 mM ATP and an ATP-regeneration system (7.5 mM phosphoenolpyruvate; 18.7 U/mL pyruvate kinase (Sigma)). GFP degradation was quantified by loss of native fluorescence (excitation 467 nm; emission 511 nm). Degradation of TAMRA-Halo-H6-ssrA or TAMRA-Halo-V13P-H6-ssrA was quantified by decreases in fluorescence (excitation 520 nm; emission 575 nm). Degradation of titinI27 unfolded by modification of cysteines with 5-iodoacetamidofluorescein (Pierce Biotechnology) was assayed by increases in fluorescence (excitation 480 nm; emission 520 nm) in the absence or presence of different concentrations of DHFR-H6-ssrA and 50 μM methotrexate (Sigma). All assays monitored by changes in fluorescence were performed using a SpectraMax M5 micro-plate reader (Molecular Devices). For degradation of TAMRA-Halo-(V13P)4-H6-ssrA monitored by SDS-PAGE, 10 μM substrate was incubated with 5 μM dye for 15 min at 30 °C before ClpXP was added. Reactions were quenched at different times by addition of SDS-loading buffer, boiled for 5 min, run on SDS gradient 4-12% polyacrylamide gels (Invitrogen), and visualized using a Typhoon-4100 imager (GE Healthcare). Band intensities were quantified using IMAGEQUANT (Molecular Dynamics).
Supplementary Material
Highlights.
ClpX pore-loop mutants translocate polypeptides at wild-type velocity against force
Single-molecule unfolding defects are quantified in optical-trap experiments
After an unfolding attempt, pore-loop variants release substrates more frequently
Pore loops function coordinate, but do not prevent, slippage between power strokes
Acknowledgements
We thank M. Aubin-Tam, M. Lang, H. Kotamarthi, K. Schmitz, and B. Stein for helpful discussions and A. Matouschek for materials. This work was supported by NIH grants GM-101988 and GM-049224. T.A.B. is an employee of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The ssrA tag is 11 residues in length. After initiation of degradation, ~35 residues of the substrate are within the ClpX pore and the ClpP chamber.
References
- Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TA, Sauer RT. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta. 2012;1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkow SR, Levchenko I, Baker TA, Sauer RT. Polypeptide translocation by the AAA+ ClpXP protease machine. Chem. Biol. 2009;16:605–612. doi: 10.1016/j.chembiol.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baytshtok V, Baker TA, Sauer RT. Assaying the kinetics of protein denaturation catalyzed by AAA+ unfolding machines and proteases. Proc. Natl. Acad. Sci. USA. 2015;112:5377–5382. doi: 10.1073/pnas.1505881112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova JC, et al. Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell. 2014;158:647–658. doi: 10.1016/j.cell.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn SE, Nager AR, Baker TA, Sauer RT. Dynamic and static components power unfolding in topologically closed rings of a AAA+ proteolytic machine. Nat. Struct. Mol. Biol. 2012;19:616–622. doi: 10.1038/nsmb.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iosefson O, Nager AR, Baker TA, Sauer RT. Coordinated gripping of substrate by subunits of an AAA+ proteolytic machine. Nat. Chem. Biol. 2015;11:201–206. doi: 10.1038/nchembio.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Sauer RT. Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc. Natl. Acad. Sci. USA. 2005;102:1390–1395. doi: 10.1073/pnas.0409634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerssemakers JW, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- Kim DY, Kim KK. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J. Biol. Chem. 2003;278:50664–50670. doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- Koodathingal P, et al. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J. Biol. Chem. 2009;284:18674–18684. doi: 10.1074/jbc.M900783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut DA, et al. Sequence- and species-dependence of proteasomal processivity. ACS Chem. Biol. 2012;7:1444–1453. doi: 10.1021/cb3001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Maillard RA, et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol. Cell. 2007;27:41–52. doi: 10.1016/j.molcel.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol. Cell. 2008a;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat. Struct. Mol. Biol. 2008b;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Protein unfolding by a AAA+ protease is dependent on ATP-hydrolysis rates and substrate energy landscapes. Nat. Struct. Mol. Biol. 2008c;15:139–145. doi: 10.1038/nsmb.1380. [DOI] [PubMed] [Google Scholar]
- Nager AR, Baker TA, Sauer RT. Stepwise unfolding of a β-barrel protein by the AAA+ ClpXP protease. J. Mol. Biol. 2011;413:4–16. doi: 10.1016/j.jmb.2011.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares AO, Nager AR, Iosefson O, Sauer RT, Baker TA. Mechanochemical basis of protein degradation by a double-ring AAA+ machine. Nat. Struct. Mol. Biol. 2014;21:871–875. doi: 10.1038/nsmb.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa I, et al. Nanomechanics of HaloTag tethers. J. Am. Chem. Soc. 2013;135:12762–12771. doi: 10.1021/ja4056382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- Sen M, et al. The ClpXP protease unfolds substrates using a constant rate of pulling but different gears. Cell. 2013;155:636–646. doi: 10.1016/j.cell.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui SM, Sauer RT, Baker TA. Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson BM, Baytshtok V, Schmitz KR, Baker TA, Sauer RT. Subunit asymmetry and roles of conformational switching in the hexameric AAA+ ring of ClpX. Nat. Struct. Mol. Biol. 2015;22:411–416. doi: 10.1038/nsmb.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Holmgren RA, Matouschek A. A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-kappaB. Nat. Struct. Mol. Biol. 2005;12:1045–1053. doi: 10.1038/nsmb1018. [DOI] [PubMed] [Google Scholar]
- Too PH, Erales J, Simen JD, Marjanovic A, Coffino P. Slippery substrates impair function of a bacterial protease ATPase by unbalancing translocation versus exit. J. Biol. Chem. 2013;288:13243–13257. doi: 10.1074/jbc.M113.452524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass RH, Chien P. Critical clamp loader processing by an essential AAA+ protease in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA. 2013;110:18138–18143. doi: 10.1073/pnas.1311302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.