Abstract
Purpose
It is known that over expression of IL6 in prostate cancer cells confer enzalutamide resistance and that this may occur through constitutive Stat3 activation. Additionally, recent pre-clinical studies suggested enzalutamide might have the potential adverse effect of inducing metastasis of prostate cancer cells via Stat3 activation. This study is aimed to target Stat3 activation and improve enzalutamide therapy.
Experimental design
Sensitivity of prostate cancer cells to enzalutamide was tested using cell growth assays and clonogenic assays. Wound healing and invasion assays were performed to determine cell migration and invasion in vitro. Quantitative reverse transcription-PCR, ELISAand Western blotting were performed to detect expression levels of PSA, c-Myc, survivin, Stat3 and AR. ChIP assay was performed to examine recruitment of AR to the PSA promoter.
Results
In the present study, we found niclosamide, a previously identified novel inhibitor of androgen receptor variant (AR-V7), inhibited Stat3 phosphorylation and expression of downstream target genes. Niclosamide synergistically reversed enzalutamide resistance in prostate cancer cells and combination treatment of niclosamide with enzalutamide significantly induced cell apoptosis and inhibited cell growth, colony formation, cell migration and invasion. Knock down of Stat3 abrogated enzalutamide resistance resulting in reduced recruitment of AR to the PSA promoter in prostate cancer cells expressing IL6. Moreover, niclosamide reversed enzalutamide resistance by down-regulating Stat3 target gene expression Stat3and abrogating recruitment of AR to PSA promoter resulting in PSA inhibition.
Conclusions
This study demonstrated the IL6-Stat3-AR axis in prostate cancer is one of the crucial mechanisms of enzalutamide resistance. Niclosamide has the potential to target the IL6-Stat3-AR pathway to overcome enzalutamide resistance and inhibit migration and invasion in advanced prostate cancer.
Keywords: prostate cancer, Stat3, interleukin-6, niclosamide, enzalutamide
Introduction
Invasion and metastasis are the fatal stage of prostate cancer patients. Enzalutamide not only prolongs the survival of men with metastatic castration-resistant prostate cancer (mCRPC) who have been pre-treated with docetaxel but also in chemo-naïve mCRPC patients (1, 2). Recent studies showed pro-metastatic effects of enzalutamide in pre-clinical models, suggesting the possibility that adverse effects of enzalutamide might present in clinical patients (3-5). Understanding the underlying mechanisms of enzalutamide resistance is urgent and would provide much needed information to clinical investigators applying early intervention strategies in patients.
In our previous study, we showed that an IL6-Stat3 feed forward loop in prostate cancer patients may be one of the crucial mechanisms involved in enzalutamide resistance (6). A recent pre-clinical study suggested that the use of enzalutamide in clinically localized prostate cancer may potentiate the formation of micro-metastases (3). Enzalutamide in prostate cancer patients might lead to unwanted side effects of enhanced macrophage infiltration / prostate cancer metastasis through modulation of CCL2-STAT3 signaling (5). Constitutively active Stat3 is a part of the positive autocrine IL6 loop and Stat3 activation in human tumors is often observed at the invasive front of tumors adjacent to inflammatory cells. This suggests that Stat3-dependent tumorigenesis is mediated by IL6 (7). Constitutive Stat3 activation in normal prostate epithelial cells enhances EMT and cell motility and active Stat3 expression was associated with tumor invasion and poor clinical outcome in patients. Due to its involvement in these events, targeting Stat3 could be an ideal strategy to inhibit prostate cancer growth and metastasis.
In the current study, we found that the IL6-Stat3-AR axis in prostate cancer patient may be one of the pivotal mechanisms of enzalutamide resistance. Niclosamide, a previously identified novel inhibitor of androgen receptor variant 7 (AR-V7)(8), was found to be able to significantly inhibit Stat3 activation and downstream target gene expression. Combining niclosamide with enzalutamide synergistically overcame enzalutamide resistance and inhibited invasion and migration in prostate cancer cells.
Materials and Methods
Reagents and Cell Culture
C4-2B, LNCaP and DU145 cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin and 0.1 mg/ml streptomycin. PZ-HPV7 cells were cultured in Keratinocyte-SFM medium supplemented with L-glutamine, epidermal growth factor (EGF) and bovine pituitary extract (BPE). LNCaP-neo and LNCaP-s17 cells were stably transfected with pcDNA3.1 vector or plasmid expressing IL6 which was described before (9), LNCaP-Stat3C cells were stably transfected with constitutively active Stat3 plasmid which was described before(10). The cells were maintained at 37°C in a humidified incubator with 5% carbon dioxide. IL6 was purchased from R&D systems. Niclosamide was purchased from Sigma-Aldrich, St. Louis, MO (N3510) and dissolved in DMSO.
Plasmids and cell transfection
For small interfering RNA (siRNA) transfection, cells were seeded at a density of 1×105 cells per well in 12-well plates and transfected with siRNA (Cell signaling #6582) targeting the Stat3 sequence; a control sequence targeting the luciferase (Luc) gene, siControl (5’-CTTACGCTGAGTACTTCGA-3’), using lipofectamine2000 (Invitrogen) at a 100 nM concentration. Cells were transiently transfected by expressing plasmids using Attractene (QIAGEN).
Luciferase Assay
LNCaP, LNCaP-s17, LNCaP-Stat3C or DU145 cells (1×105 cells per well of 12-well plate) were transfected with 0.5 μg of pLucTKS3 reporter plasmid containing specific responsive elements for Stat3 or the control plasmid. The luciferase activity was determined 24 –48 hr after transfection using a dual-luciferase reporter assay system (Promega).
Chromatin immunoprecipitation assay
LNCaP-s17 cells were treated as indicated. DNA-AR protein complexes were cross-linked inside the cells by the addition of 1% formaldehyde. Whole-cell extracts were prepared by sonication, and an aliquot of the cross-linked DNA-protein complexes was immunoprecipitated by incubation with the AR-specific antibody (AR-C19; Santa Cruz Biotechnology) overnight at 4°C with rotation. Chromatin-antibody complexes were isolated from solution by incubation with protein A/G agarose beads for 1 hour at 4°C with rotation. The bound DNA-protein complexes were washed and eluted from beads with elution buffer (1% SDS and 0.1 mol/L NaHCO3), cross links were reversed, and DNA was extracted. The resulting chromatin preparations were analyzed by PCR using primers spanning either the proximal or the distal enhancer AREs of the PSA promoter. Primers used for ChIP assay were: ARE: 5’-CCTAGATGAAGTCTCCATGAGCTACA, ARE 3’-GGGAGGGAGAGCTAGCACTTG. Isotype-matched IgG was used as control (11).
Western blot analysis
Whole cell protein extracts were resolved on SDS–PAGE and proteins were transferred to nitrocellulose membranes. After blocking for 1 hour at room temperature in 5% milk in PBS/0.1% Tween-20, membranes were incubated overnight at 4°C with the indicated primary antibodies. p-Stat3(Tyr705) (sc-7993-R, rabbit polyclonal antibody, 1:1000 dilution), Stat3 (sc-482, rabbit polyclonal antibody, 1:1000 dilution), AR (441, sc-7305, Mouse monoclonal antibody, 1:1000 dilution) and c-Myc (sc-764, rabbit polyclonal antibody, 1:1000 dilution) were purchased from Santa Cruz Biotechnology, Santa Cruz, CA; Survivin (FL-142, rabbit polyclonal antibody, 1:1000 dilution) and Tubulin (T5168, Monoclonal Anti-α-Tubulin antibody, 1:5000 dilution) were purchased from Sigma-Aldrich, St. Louis, MO. Tubulin was used to monitor the amounts of samples applied. Following secondary antibody incubation, immunoreactive proteins were visualized with an enhanced chemiluminescence detection system (Millipore, Billerica, MA).
Cell growth assay
LNCaP-s17 cells were seeded in 12-well plates at a density of 1×105 cells/well in RPMI 1640 media containing 10% FBS and treated with 0.25 or 0.5 μM niclosamide with or without 20 μM enzalutamide for 48 hours and total cell number was counted.
Clonogenic Assay
LNCaP-s17 cells or LNCaP-Stat3C stable clone cells were treated with DMSO, enzalutamide, niclosamide or combination in FBS conditions. Cells were plated at equal density (1000 cells/dish) in 100 mm dishes for 14 days. The cells were rinsed with PBS before staining with 0.5% crystal violet/4% formaldehyde for 30 min and the number of colonies was counted (12, 13).
Cell death ELISA
LNCaP-s17 cells were seeded on 12-well plates (1 × 105 cells/well) in RPMI 1640 media containing 10% FBS and treated with 0.25 or 0.5 μM niclosamide with or without 20 μM enzalutamide for 48 hours. Mono- and oligonucleosomes in the cytoplasmic fraction were measured by the Cell Death Detection ELISA kit (Roche, Cat. NO. 11544675001) according to the manufacturer's instructions as described before (12, 14). Briefly, floating and attached cells were collected and homogenized in 400 μL of incubation buffer. The wells were coated with antihistone antibodies and incubated with the lysates, horseradish peroxidase–conjugated anti-DNA antibodies, and the substrate, and absorbance was read at 405 nm.
Wound Healing Assay
DU145, LNCaP-s17 and LNCaP-Stat3C cells were cultured under standard condition as described above and plated onto 60 mm2 dishes. Scratch wounds were introduced into the confluent monolayers the day after plating. The wounds were created with a 200 μL pipette tip. Cells were kept in regular culture media with various concentrations of niclosamide or combined with enzalutamide. Wound closure was monitored over time and photographed using Olympus 1 × 81 microscope at 40× magnification. The wound closure was quantified by measuring the remaining unmigrated area using ImageJ software.
Invasion Assay
LNCaP-Stat3C or DU145 cells (1×104) were suspended in serum free RPMI1640 medium containing niclosamide or enzalutamide into the cell culture inserts for 24-well plates containing 8 mm pores. The inserts were coated with Matrigel (2-3 mg/ml protein) and allowed to solidify for 2 hours before cell plating. The lower chambers were filled with 300 μl growth medium containing DMSO or drug as indicated. After 48 h, cells were fixed with 5% glutaraldehyde in PBS, washed with PBS, and stained with 0.5% toluidine blue in 2% Na2CO3 solution. Invasive cells that penetrated the membrane were counted under the microscope, each group was counted at least 3 visual areas (random 40×high-power fields).
Measurement of PSA
PSA levels were measured in the culture supernatants using ELISA (United Biotech, Inc., Mountain View, CA) according to the manufacturer's instructions as described before (15).
Statistical Analysis
All data are presented as means ± standard deviation of the mean (SD). Statistical analyses were performed with Microsoft Excel analysis tools. Differences between individual groups were analyzed by one-way analysis of variance (ANOVA) followed by the Scheffé procedure for comparison of means. P < 0.05 was considered statistically significant.
Results
Niclosamide inhibited Stat3 phosphorylation and downstream target genes expression in prostate cancer cells
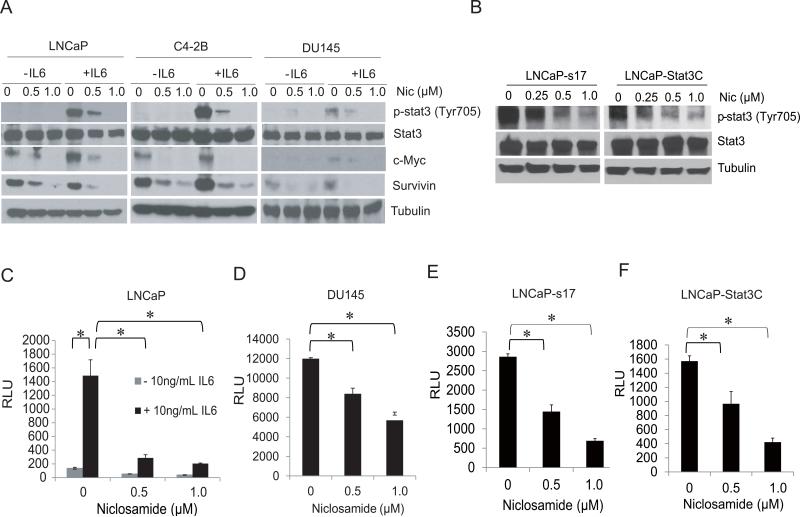
Niclosamide has been shown to inhibit Stat3 in colon and head-neck cancer cells (16, 17). To determine if niclosamide also inhibits Stat3 activation in prostate cancer cells, LNCaP, C4-2B or DU145 cells were treated with different doses of niclosamide overnight and then stimulated with 10ng/ml IL6 for 30 minutes. As shown in Fig.1A, niclosamide significantly inhibited IL6 induced Stat3 phosphorylation in these cell lines. Notably, niclosamide inhibited both endogenous c-Myc and survivin protein expression as well as expression induced by IL6. Our previous data showed LNCaP-s17 cells and LNCaP-Stat3C cells which stably express IL6 and have constitutive Stat3 activation respectively (18). To examine whether niclosamide inhibits endogenous Stat3 activation, LNCaP-s17 and LNCaP-Stat3C cells were treated with different concentrations niclosamide overnight and Stat3 phosphorylation was examined. As shown in Fig.1B, niclosamide significantly inhibited Stat3 phosphorylation (Tyr705) in a dose dependent manner. To examine the effect of niclosamide on the activity of Stat3-responsive genes, we transfected LNCaP, DU145, LNCaP-s17 and LNCaP-Stat3C cells with the pLucTKS3 luciferase reporter containing the Stat3 responsive elements or control plasmids and treated the cells with niclosamide in the presence or absence of IL6. As shown in Figure 1C, IL6 induced Stat3-responsive luciferase reporter activity in LNCaP cells, which was reduced by niclosamide treatment. DU145, LNCaP-s17 and LNCaP-Stat3C cells exhibited constitutive activation of Stat3. Niclosamide also decreased the Stat3-responsive luciferase activity in a dose-dependent manner (Fig.1D-1F). Collectively, these data suggest that niclosamide inhibits both IL6-induced and constitutive Stat3 activation and Stat3 mediated gene expression.
Figure 1. Niclosamide inhibited Stat3 activation in prostate cancer cells.
A) LNCaP, C4-2B or DU145 cells were treated with DMSO, 0.5 μM or 1 μM niclosamide overnight, cells were then stimulated with 10 ng/mL of IL6 for 30 minutes. Whole cell lysates were prepared and subjected to Western blot analysis using antibodies as indicated. B) Niclosamide inhibited endogenous Stat3 activation. LNCaP-s17 and LNCaP-Stat3C cells were treated with DMSO, 0.5 μM, or 1 μM niclosamide overnight, Whole cell lysates were prepared and subjected to Western blot analysis using antibodies as indicated. C) Niclosamide inhibited Stat3 activity in LNCaP cells. LNCaP cells were transient transfected with 0.5 μg of pLucTKS3 reporter plasmid containing specific responsive elements for Stat3 or the control plasmid with or without 10 ng/mL IL6 for 24 hours. Cells were then treated with 0.5 μM or 1 μM niclosamide overnight and Luciferase activity was measured. D-F) DU145, LNCaP-s17 or LNCaP-Stat3C cells were transiently transfected with 0.5 μg of pLucTKS3 reporter plasmid containing specific responsive elements for Stat3 or the control plasmid for 24 hours, cells were then treated with 0.5 μM or 1 μM niclosamide overnight and Luciferase activity was measured. Results are presented as means ± SD of 3 experiments performed in duplicate. * P<0.05 Nic: Niclosamide.
Niclosamide inhibited cell invasion and colony formation in prostate cancer cells
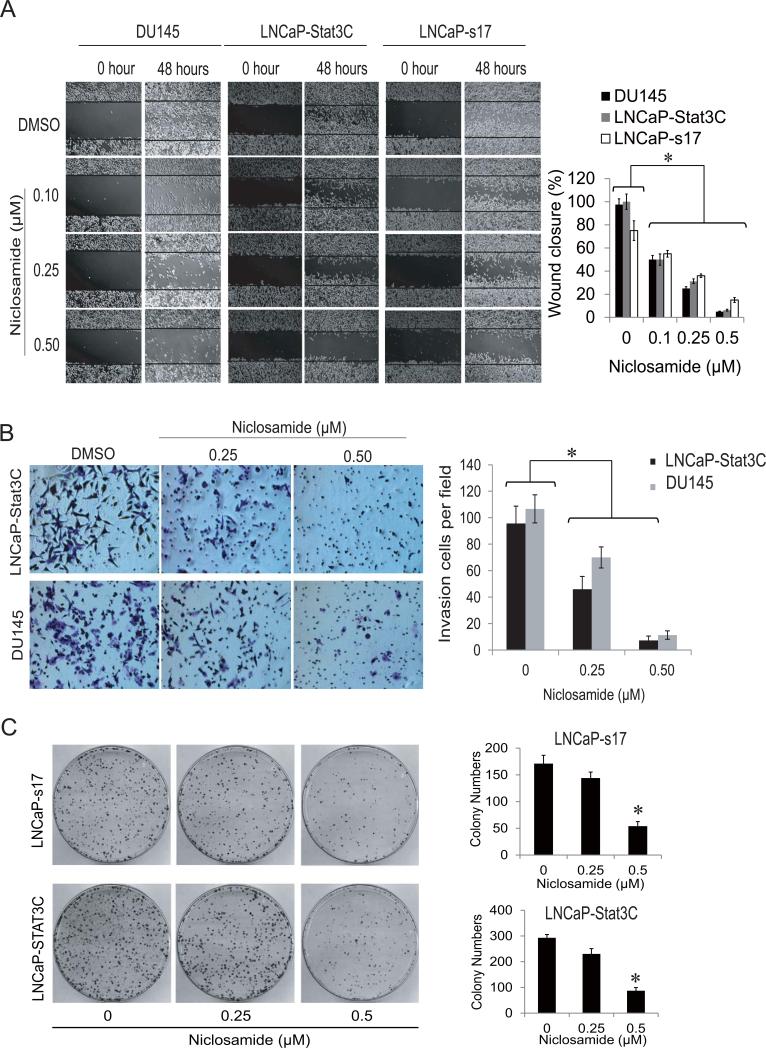
Evidence suggests constitutive Stat3 activation is oncogenic and contributes to tumor progression and metastasis (19-21). To test whether niclosamide inhibits cell migration and invasion, wound healing assays were performed in Stat3 constitutively activated LNCaP-Stat3C, LNCaP-s17 and DU145 cells. As shown in Fig.2A, niclosamide inhibited wound healing in a dose dependent manner in each of these cell lines which express constitutively active Stat3. To further investigate if LNCaP-Stat3C and DU145 cells have higher migration ability, a Boyden chamber based invasion assay were performed on these two cell lines. Niclosamide significantly reduced the number of invasive cells in a dose dependent manner in both cell lines (Fig.2B). Previously, we have shown niclosamide inhibited colony formation ability in AR-V7 overexpressing prostate cancer cells (8). To test if niclosamide also has the ability to inhibit colony formation in constitutively active Stat3 prostate cancer cells, LNCaP-s17 and LNCaPStat3C cells were treated with 0.25 μM or 0.5 μM niclosamide. As depicted in Fig.2C, 0.25 μM niclosamide slightly inhibited colony formation while 0.5 μM niclosamide significantly reduced colony number and size in both cell lines. These data showed that niclosamide inhibits cell invasion and colony formation in prostate cancer cells.
Figure 2. Niclosamide inhibited cell migration, invasion and colony formation of prostate cancer cells.
A) DU145, LNCaP-S17 or LNCaP-Stat3C cells were treated with 0.1μM, 0.25 μM or 0.5 μM niclosamide in media containing FBS after wound were introduced, photographs of the cells were taken and wound closure was quantified at the time indicated. B) Representative photographs of invasive LNCaP-Stat3C and DU145 cells. The cells were allowed to migrate through matrigel coated membranes with 8 mm pores for 48 hours in the presence of 0.1μM, 0.25 μM or 0.5 μM niclosamide. Invasive cells were counted under microscope (right panel). C) Colony formation was examined by clonogenic assay. LNCaP-s17 and LNCaP-Stat3C cells were treated with 0.25 μM or 0.5 μM niclosamide and equal amounts of cells were plated in 10cm dishes. The colonies were stained with 0.5% crystal violet after 3 weeks, and colony number was counted. Results are presented as means ± SD of 3 experiments performed in triplicate. * P < 0.05
Niclosamide synergistically enhanced enzalutamide treatment in constitutively active Stat3 prostate cancer cells
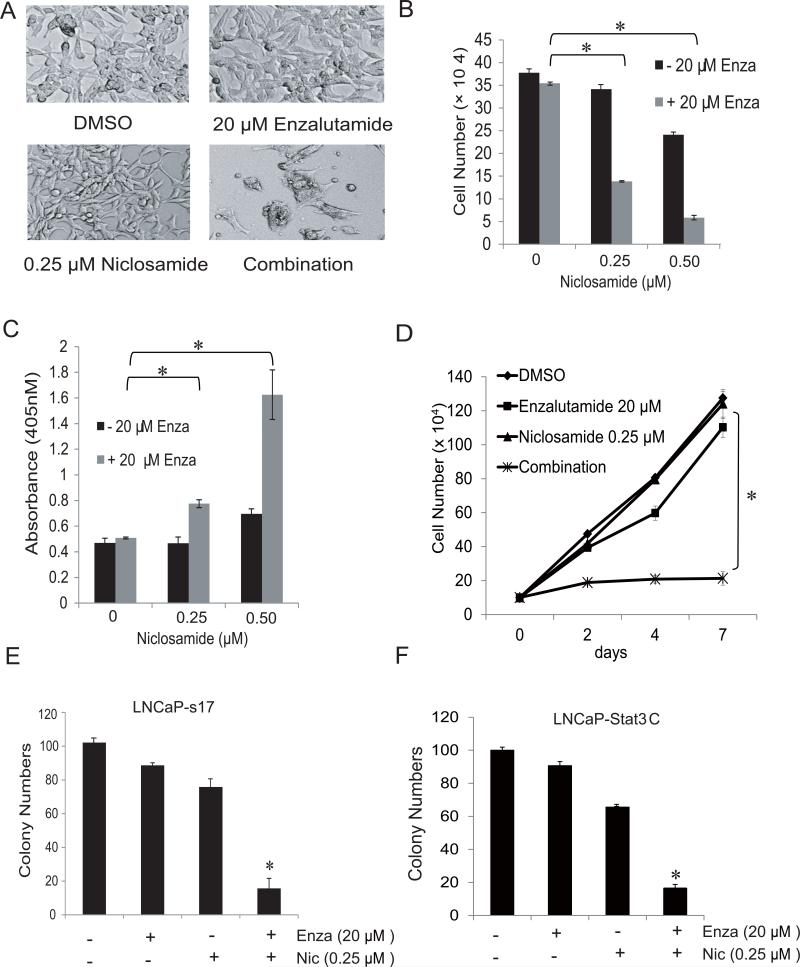
In our previous study, we observed that niclosamide synergistically enhanced enzalutamide treatment in CWR22Rv1 and C4-2B MDVR cells through AR variant inhibition (8). We next examined the combinatory effects of enzalutamide and niclosamide in constitutively active Stat3 prostate cancer cells. As shown in Fig3A, neither 20μM enzalutamide nor 0.25 μM niclosamide alone changed cell morphology of LNCaP-s17 cells. Conversely, in combination the two drugs dramatically inhibited cell growth and modified cell morphology. To further examine the combinatory effects of these two drugs, LNCaP-s17 cells were treated with two different concentration of niclosamide (0.25 and 0.5μM) combined with 20μM enzalutamide in. After 48 hours, cell numbers were counted and supernatant was collected for cell death detection. As depicted in Fig.3B-C, niclosamide combined with enzalutamide significantly inhibited cell growth and induced cell death of LNCaP-s17 cells. These results were also observed in a time dependent experiment; LNCaP-s17 cells were treated with 20μM enzalutamide together with 0.25μM niclosamide for different amounts of time. The growth of cells treated with enzalutamide or niclosamide alone continued during the 7 days treatment while growth of cells in the co-treatment group was completely inhibited (Fig.3D). To further test the combination treatment on cell function change, a clonogenic assay was performed. As shown in Fig.3E, enzalutamide combined with niclosamide significantly inhibited LNCaP-s17 colony formation. We confirmed these results in LNCaP-Stat3C cells (Fig.3F). Taken together, the results above showed niclosamide synergistically enhanced enzalutamide treatment in constitutive active Stat3 prostate cancer cells.
Figure 3. Niclosamide synergistically reversed enzalutamide resistance in prostate cancer cells.
A) Representative pictures of cell morphology changes in LNCaP-s17 cells after treatment with different regimens as indicated. LNCaP-s17 cells were treated with 0.25 μM or 0.5 μM niclosamide with or without 20 μM enzalutamide in media containing FBS. Cell numbers were counted after 48 h (B) and apoptosis was analyzed by Cell death ELISA (C). Results are presented as means ± SD of 3 experiments performed in duplicate. D) LNCaP-s17 cells were treated with 0.25 μM niclosamide with or without 20 μM enzalutamide in media containing FBS and cell numbers were counted after 2, 4 and 7 days. Results are presented as means ± SD of 3 experiments performed in duplicate. E) Colony formation was examined by clonogenic assay. LNCaP-s17 cells were treated with 0.25 μM niclosamide with or without 20 μM enzalutamide and equal amount cells were plated in 10cm dishes. The colonies were stained with 0.5% crystal violet after 3 weeks and colonies numbers were counted. F) LNCaP-Stat3C cells were treated with 0.25 μM niclosamide with or without 20 μM enzalutamide and equal amount cells were plated in 10cm dishes. The colonies were stained with 0.5% crystal violet after 3 weeks and colony number was counted. Results are presented as means ± SD of 3 experiments performed in duplicate.* P< 0.05 Enza: Enzalutamide, Nic: Niclosamide.
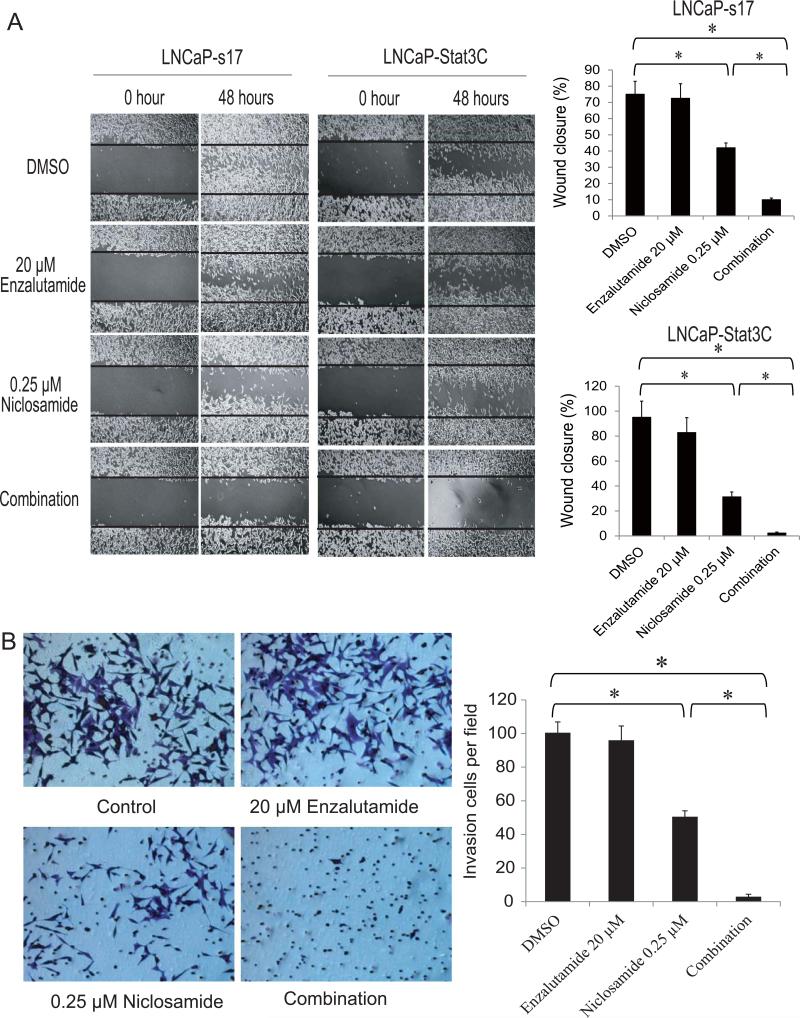
Several reports have shown that activated Stat3 is linked to prostate cancer recurrence and metastasis (21, 22). To continue to test if the combinatory treatment of enzalutamide with niclosamide affected cell migration and invasion, a wound healing assay was performed in LNCaP-s17 cells and LNCaP-Stat3C cells. Alone, 20μM enzalutamide had little effect on cell wound healing in these two cell lines. However, when combined with 0.25μM niclosamide, wound healing was totally inhibited (Fig.4A). We also performed Boyden chamber based invasion assays in LNCaP-Stat3C cells. Twenty micromolar enzalutamide did not inhibit LNCaP-Stat3C invasion, 0.25μM niclosamide had a 50% reduction of invasive cells, while the combination treatment reduced more than 90% of invasive cells (Fig.4B). Collectively, these data suggested that by acting as a potent Stat3 inhibitor, niclosamide synergistically enhanced enzalutamide effects by inhibiting cell migration and invasion in constitutively active Stat3 prostate cancer cells.
Figure 4. Co-treatment with enzalutamide and niclosamide inhibit prostate cancer cell migration and invasion.
A) LNCaP-s17 and LNCaP-Stat3C cells were treated with 0.25 μM niclosamide with or without 20 μM enzalutamide in media containing FBS. After wounds were introduced, photographs of the cells were taken and wound closure was quantified at the time indicated. B) Representative photographs of invasive LNCaP-Stat3C cells. The cells were allowed to migrate through matrigel coated membranes with 8 μm pores for 48 hours in the presence of 0.25 μM niclosamide with or without 20 μM enzalutamide. Invasive LNCaP-Stat3C cells were counted under microscope (right panel). Results are presented as means ± SD of 3 experiments performed in triplicate. * P< 0.05
Stat3 inhibition enhanced enzalutamide treatment by targeting AR signaling
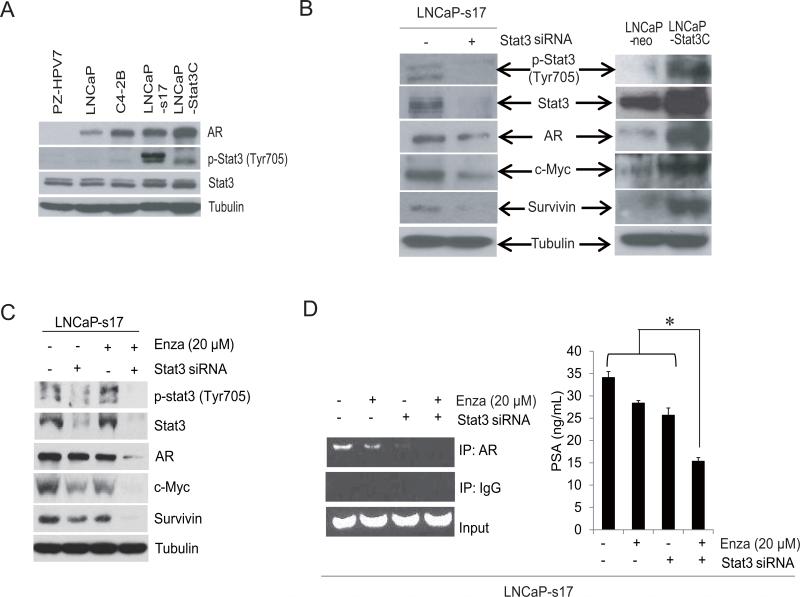
In our previous study, we revealed that the IL6-Stat3 feed forward loop might be one of the important pathways contributing to enzalutamide resistance (6). To further explore the underlying mechanisms, AR and Stat3 activation were examined in different cell lines. As shown in Fig.5A, the normal prostate epithelial cell line PZ-HPV7 does not express AR nor activated Stat3. C4-2B, LNCaP-s17 and LNCaP-Stat3C cells expressed higher AR than LNCaP parental cells. Additionally, LNCaP-s17 and LNCaP-Stat3C cells expressed constitutively active Stat3. To examine genes differentially expressed in response to constitutively active Stat3, LNCaP-s17 cells were transiently transfected with Stat3 siRNA. Knock down of Stat3 in these cells significantly inhibited expression of c-Myc and survivin, and also partially inhibited AR expression (Fig.5B). On the other hand, constitutive activation of Stat3 in LNCaP cells significantly enhanced expression of these genes.
Figure 5. Stat3 activation induced enzalutamide resistance linked to the recruitment of AR to the PSA promoter.
A) LNCaP, C4-2B, LNCaP-s17, LNCaP-Stat3C or PZ-HPV7 cell protein was harvested and immunoblotted with the antibody indicated. B) LNCaP-s17 cells were transfected with siRNA specific to Stat3 or control siRNA and whole cell lysate was extracted and immunoblotted with antibody indicated. Whole cell lysate from LNCaP-neo and LNCaPStat3C cells was extracted and immunoblotted with the antibody indicated. C) LNCaP-s17 cells were transfected with siRNA specific to Stat3 or control siRNA and were treated with or without 20 μM enzalutamide for 2 days and whole cell protein was extracted and immunoblotted with the antibody indicated. D) Recruitment of AR to AREs in the PSA promoter was analyzed by ChIP assay, LNCaP-s17 cells was transfected with siRNA specific to Stat3 or control siRNA and were treated with or without 20 μM enzalutamide overnight. Recruitment of AR to AREs in the PSA promoter was analyzed by ChIP assay (left) and the supernatant was collected for PSA ELISA analysis (right). * P< 0.05 Enza: Enzalutamide.
To determine whether knock down of Stat3 enhanced inhibition of Stat3 target genes induced by enzalutamide, western blots were performed. Knock down of Stat3 significantly enhanced the effects of enzalutamide treatment and inhibited expression of c-Myc, survivin and AR (Fig.5C). To further confirm that the Stat3-AR axis is involved in enzalutamide resistance, a ChIP assay was performed. As shown in Fig.5D left, enzalutamide slightly inhibited AR recruitment to the PSA promoter in LNCaP-s17 cells and knock down of Stat3 significantly reduced recruitment of AR to the PSA binding site. Enzalutamide combined with Stat3 siRNA totally blocked AR recruitment to the PSA promoter. To examine if the recruitment of AR to AREs translated to AR activation, PSA ELISA was performed. As shown in the right panel of Fig.5D, enzalutamide combined with Stat3 siRNA significantly inhibited PSA expression in LNCaP-s17 cells. These results suggested that enzalutamide resistance induced by constitutive Stat3 activation is due to higher recruitment of AR to the PSA promoter. Targeting the Stat3-AR axis is a promising strategy to overcome enzalutamide resistance.
Niclosamide inhibited the recruitment of AR to the PSA promoter by blocking constitutively active Stat3
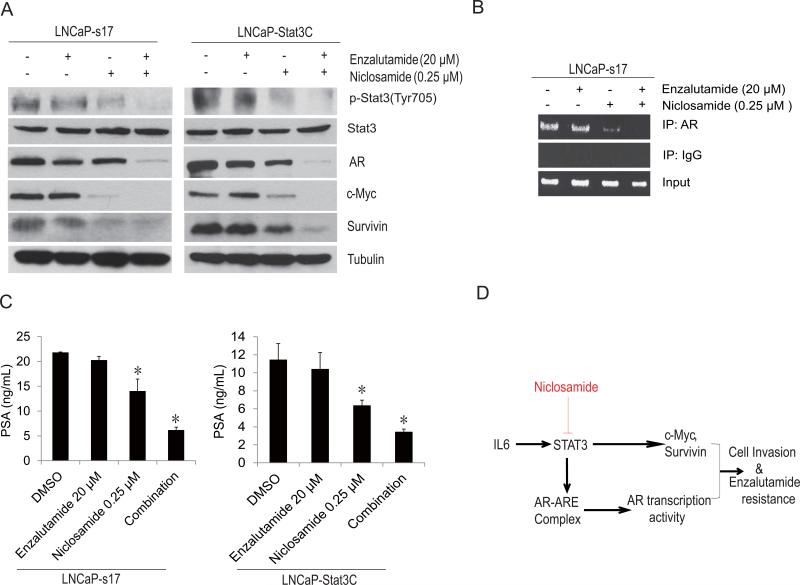
The studies suggest that niclosamide functions not only as an AR variant inhibitor but also as an inhibitor of Stat3 activation. To further address the mechanisms of the combinatory effects of niclosamide with enzalutamide, Stat3 downstream target genes were examined. LNCaP-s17 and LNCaP-Stat3C cells were treated with different regimens, as indicated in Fig.6A. Combination treatment significantly inhibited Stat3 phosphorylation and expression of survivin and c-Myc compared to niclosamide or enzalutamide alone. AR expression was also inhibited in the combination group.
Figure 6. Co-treatment with enzalutamide and niclosamide down regulate Stat3 target gene expression and inhibit AR transcriptional activity.
A) LNCaP-s17 and LNCaP-Stat3C cells were treated with 0.25 μM niclosamide with or without 20 μM enzalutamide in media containing FBS for 48 hours. Whole cell protein was extracted and immunoblotted with the antibody indicated. B) LNCaP-s17 cells were treated with 0.25 μM niclosamide with or without 20 μM enzalutamide overnight. Recruitment of AR to AREs in the PSA promoter was analyzed by ChIP assay. C) The supernatant was collected for PSA ELISA analysis. Results are presented as means ± SD of 3 experiments performed in triplicate. D) Proposed pathway of enzalutamide resistance in constitutively active Stat3 prostate cancer cells. * P< 0.05
To examine whether co-treatment also affected the AR recruitment, a ChIP assay was performed; LNCaP-s17 cells were treated with DMSO, 20μM enzalutamide or 0.25μM niclosamide alone or in combination overnight. As shown in Fig.6B, treatment with enzalutamide alone was not able to block AR recruitment to the PSA binding site, while niclosamide alone blocked AR recruitment to the PSA binding site. However, combination treatment with enzalutamide further inhibited AR recruitment to the PSA promoter. To test if the recruitment of AR also affected AR activity, PSA ELISA was performed. As depicted in Fig.6C, treatment with 0.25 μM niclosamide slightly down regulated PSA secretion in both LNCaP-s17 and LNCaP-Stat3C cells while niclosamide/enzalutamide co-treatment significantly reduced PSA in both cell lines by an additional 50%. Collectively, these data showed that the IL6-Stat3-AR axis (Fig.6D) is involved in enzalutamide resistance and that niclosamide reversed enzalutamide resistance and inhibited cell invasion and migration in prostate cancer by inhibiting Stat3.
Discussion
For prostate cancer patients, metastasis is the primary underlying cause of fatality. For men with the metastatic prostate cancer, only one-third survive for five years after diagnosis (23). The molecular mechanisms of metastasis in prostate cancer are still under intense investigation. To date, several mechanisms have been identified, including cell-matrix adhesion, matrix degradation and cytokine activation (24, 25). Recently, data has suggested that androgen deprivation therapy (ADT) might induce prostate cancer metastasis. It has been noted that the second generation anti-androgen enzalutamide, which significantly increased survival benefit in mCRPC patients, may enhance metastasis in pre-clinical models. Lin et.al (5)observed that enzalutamide promoted prostate cancer metastasis via enhancing macrophage infiltration in both in vitro co-culture systems and in vivo mouse models. They proposed the underlying mechanism might involve CCL2/Stat3 signaling and suggested that a new combination therapy to suppress prostate cancer proliferation with the ADT in addition to a new therapy to overcome the metastasis could be beneficial. Asangani et.al (3) showed that in a VCaP xenograft mouse model, enzalutamide treatment significantly enhanced liver and femur metastasis, and suggested that the use of enzalutamide in clinically localized prostate cancer may potentiate the formation of micro-metastases. Identifying new therapies to suppress the potential metastatic side effects and to overcome enzalutamide resistance is critical.
Our previous study reported that IL6 overexpression enhanced AR-ARE DNA binding activity and increased AR nuclear translocation in LNCaP cells (18). IL6 may regulate intraprostatic androgen synthesis in the absence of exogenous steroid precursors through regulating steroidogenic enzyme gene expression and inducing neuroendocrine differentiation (NED) through suppression of RE-1 silencing transcription factor (REST) (26, 27). Autocrine IL6 in prostate cancer cells significantly induced enzalutamide resistance, indicating that serum IL6 levels might be one of the pivotal factors in enzalutamide treatment and up regulation of IL6 in the patients’ serum may be one of the negative therapeutic markers associated with enzalutamide. We found that the IL6-Stat3 feed forward loop was involved in enzalutamide resistance and targeting Stat3 could be a promising strategy to overcome this resistance (6). In the present study, we determined that constitutively active Stat3 in prostate cancer cells activated downstream expression of c-Myc and survivin, and knock down of Stat3 in LNCaP-s17 cells significantly abrogated AR recruitment to ARE binding sites and resensitized the cells to enzalutamide treatment. Of note, knock down Stat3 also resultant target genes such as c-Myc and survivin inhibition might relate to cell migration and invasion. Overexpression of Myc has been linked to genomic instability, changes in morphology and function such as epithelialmesenchymal transition (EMT) and metastasis which might through miRNA regulation and Bmi-1 activation (28). Survivin has been associated with increased invasion and metastasis of colorectal cancer (29) hepatocellular carcinoma (30) and breast cancer (31). Taken together, these data suggested the IL6-Stat3-AR axis is a crucial mechanism involved in enzalutamide resistance. Target the Stat3 which is the upstream transcriptional regulator of c-Myc and survivin is a promising strategy to suppress the cell migration and invasion in prostate cancer.
Constitutive Stat3 activation has been observed in many advanced cancers and is associated with poor prognosis(32). Constitutively active Stat3 is accompanied by high levels of IL6 expression, indicating IL6 may be the main driver of Stat3 persistent activation and blocking the IL6-Stat3 axis could be a potent strategy to reverse enzalutamide resistance(33). Niclosamide is a FDA approved drug which has been used to treat tapeworm infection for about 50 years(34). In our previous study, we identified niclosamide as a novel AR variant inhibitor in prostate cancer cells and observed that it reversed enzalutamide resistance dramatically (8). In the current study, we demonstrated that niclosamide is also a potent Stat3 inhibitor in prostate cancer cells and found that it significantly reversed enzalutamide resistance in constitutively active Stat3 prostate cancer cells. Co-treatment with enzalutamide and niclosamide not only inhibited cell proliferation but also inhibited cell invasion. Stat3 activation has been found to be associated with lymph node and bone metastases of clinical human prostate cancer (21, 22). In our present study, we observed that enzalutamide alone cannot inhibit constitutively activated Stat3-mediated prostate cancer cell invasion. However, when combined with niclosamide, the number of invasive cells was significantly reduced. Co-treatment not only down regulated Stat3 target genes, but also reduced recruitment of AR to the PSA promoter. The interesting finding in this study is combination treatment dramatically reduced AR protein level in prostate cancer cells. Our previous study suggested niclosamide down regulated AR-V7 level by protein degradation might through a proteasome dependent pathway(8). Enzalutamide induce AR protein degradation by facilitating SPOP-AR interaction (35), SPOP is a bona fide E3 ligase of the AR, it is possible that combination treatment of niclosamide and enzalutamide could also enhance AR protein degradation by promoting the functional SPOP bind to the consensus motif of AR hinge domain. Our previous study demonstrated overexpressed IL6 resultant constitutive active Stat3 in prostate cancer cells may counteract enzalutamide action in blocking the recruitment of AR to target gene promoters (6), Stat3 has been shown to interact with AR N terminal domain (NTD) and activate AR transcription (36). Inhibition Stat3 activation by niclosamide might block the AR recruitment driven by Stat3 and combination treatment of niclosamide and enzalutamide down regulate AR protein level also resultant the inhibition of AR recruitment to PSA promoter.
In summary, the IL6-Stat3-AR axis is one of the crucial mechanisms contributing to enzalutamide resistance. Targeting Stat3 is a promising strategy to both enhance enzalutamide treatment and reverse adverse side effects associated with enzalutamide. Our studies demonstrate that niclosamide inhibits constitutively activated Stat3 and AR variants and suppresses cell proliferation, migration and invasion. Notably, niclosamide has the ability to synergistically enhance the anticancer effects of enzalutamide in advanced prostate cancer cells. Targeting Stat3 and AR variants with niclosamide holds the promise for treatment of advanced prostate cancer.
Acknowledgements
This work is supported in part by grants NIH/NCI CA140468, CA168601, CA179970 (A.C. Gao), US Department of Veterans Affairs, Office of Research and Development VA Merits I01 BX000526 (A.C. Gao), and by resources from the VA Northern California Health Care System, Sacramento, California.
References
- 1.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–97. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 2.Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. The New England journal of medicine. 2014;371:424–33. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–82. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin TH, Lee SO, Niu Y, Xu D, Liang L, Li L, et al. Differential androgen deprivation therapies with anti-androgens casodex/bicalutamide or MDV3100/Enzalutamide versus anti-androgen receptor ASCJ9(R) Lead to promotion versus suppression of prostate cancer metastasis. The Journal of biological chemistry. 2013;288:19359–69. doi: 10.1074/jbc.M113.477216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin TH, Izumi K, Lee SO, Lin WJ, Yeh S, Chang C. Anti-androgen receptor ASC-J9 versus anti-androgens MDV3100 (Enzalutamide) or Casodex (Bicalutamide) leads to opposite effects on prostate cancer metastasis via differential modulation of macrophage infiltration and STAT3-CCL2 signaling. Cell death & disease. 2013;4:e764. doi: 10.1038/cddis.2013.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C, Zhu Y, Lou W, Cui Y, Evans CP, Gao AC. Inhibition of constitutively active Stat3 reverses enzalutamide resistance in LNCaP derivative prostate cancer cells. The Prostate. 2014;74:201–9. doi: 10.1002/pros.22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bromberg J, Wang TC. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell. 2009;15:79–80. doi: 10.1016/j.ccr.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu C, Lou W, Zhu Y, Nadiminty N, Schwartz CT, Evans CP, et al. Niclosamide Inhibits Androgen Receptor Variants Expression and Overcomes Enzalutamide Resistance in Castration-Resistant Prostate Cancer. Clinical Cancer Research. 2014;20:3198–210. doi: 10.1158/1078-0432.CCR-13-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lou W, Ni Z, Dyer K, Tweardy DJ, Gao AC. Interleukin-6 induces prostate cancer cell growth accompanied by activation of stat3 signaling pathway. The Prostate. 2000;42:239–42. doi: 10.1002/(sici)1097-0045(20000215)42:3<239::aid-pros10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 10.DeMiguel F, Lee SO, Lou W, Xiao X, Pflug BR, Nelson JB, et al. Stat3 enhances the growth of LNCaP human prostate cancer cells in intact and castrated male nude mice. The Prostate. 2002;52:123–9. doi: 10.1002/pros.10110. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Nadiminty N, Tummala R, Chun JY, Lou W, Zhu Y, et al. Andrographolide targets androgen receptor pathway in castration-resistant prostate cancer. Genes & cancer. 2011;2:151–9. doi: 10.1177/1947601911409744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu C, Zhu Y, Lou W, Nadiminty N, Chen X, Zhou Q, et al. Functional p53 determines docetaxel sensitivity in prostate cancer cells. The Prostate. 2013;73:418–27. doi: 10.1002/pros.22583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C, Lou W, Zhu Y, Yang JC, Nadiminty N, Gaikwad NW, et al. Intracrine Androgens and AKR1C3 Activation Confer Resistance to Enzalutamide in Prostate Cancer. Cancer research. 2015 Feb 3; doi: 10.1158/0008-5472.CAN-14-3080. pii: canres.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, Liu C, Nadiminty N, Lou W, Tummala R, Evans CP, et al. Inhibition of ABCB1 expression overcomes acquired docetaxel resistance in prostate cancer. Molecular cancer therapeutics. 2013;12:1829–36. doi: 10.1158/1535-7163.MCT-13-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui Y, Nadiminty N, Liu C, Lou W, Schwartz CT, Gao AC. Upregulation of glucose metabolism by NF-kappaB2/p52 mediates enzalutamide resistance in castration-resistant prostate cancer cells. Endocrine-related cancer. 2014;21:435–42. doi: 10.1530/ERC-14-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li R, Hu Z, Sun SY, Chen ZG, Owonikoko TK, Sica GL, et al. Niclosamide overcomes acquired resistance to erlotinib through suppression of STAT3 in non-small cell lung cancer. Molecular cancer therapeutics. 2013;12:2200–12. doi: 10.1158/1535-7163.MCT-13-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Safdari Y, Khalili M, Farajnia S, Asgharzadeh M, Yazdani Y, Sadeghi M. Recent advances in head and neck squamous cell carcinoma--a review. Clinical biochemistry. 2014;47:1195–202. doi: 10.1016/j.clinbiochem.2014.05.066. [DOI] [PubMed] [Google Scholar]
- 18.Lee SO, Lou W, Hou M, de Miguel F, Gerber L, Gao AC. Interleukin-6 promotes androgen-independent growth in LNCaP human prostate cancer cells. Clin Cancer Res. 2003;9:370–6. [PubMed] [Google Scholar]
- 19.Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–97. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 21.Abdulghani J, Gu L, Dagvadorj A, Lutz J, Leiby B, Bonuccelli G, et al. Stat3 promotes metastatic progression of prostate cancer. Am J Pathol. 2008;172:1717–28. doi: 10.2353/ajpath.2008.071054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun M, Liu C, Nadiminty N, Lou W, Zhu Y, Yang J, et al. Inhibition of Stat3 activation by sanguinarine suppresses prostate cancer cell growth and invasion. The Prostate. 72:82–9. doi: 10.1002/pros.21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norgaard M, Jensen AO, Jacobsen JB, Cetin K, Fryzek JP, Sorensen HT. Skeletal related events, bone metastasis and survival of prostate cancer: a population based cohort study in Denmark (1999 to 2007). The Journal of urology. 2010;184:162–7. doi: 10.1016/j.juro.2010.03.034. [DOI] [PubMed] [Google Scholar]
- 24.Holzapfel BM, Wagner F, Loessner D, Holzapfel NP, Thibaudeau L, Crawford R, et al. Species-specific homing mechanisms of human prostate cancer metastasis in tissue engineered bone. Biomaterials. 2014;35:4108–15. doi: 10.1016/j.biomaterials.2014.01.062. [DOI] [PubMed] [Google Scholar]
- 25.Roodman GD. Mechanisms of bone metastasis. N Engl J Med. 2004;350:1655–64. doi: 10.1056/NEJMra030831. [DOI] [PubMed] [Google Scholar]
- 26.Chun JY, Nadiminty N, Dutt S, Lou W, Yang JC, Kung HJ, et al. Interleukin-6 regulates androgen synthesis in prostate cancer cells. Clin Cancer Res. 2009;15:4815–22. doi: 10.1158/1078-0432.CCR-09-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Y, Liu C, Cui Y, Nadiminty N, Lou W, Gao AC. Interleukin-6 induces neuroendocrine differentiation (NED) through suppression of RE-1 silencing transcription factor (REST). The Prostate. 2014;74:1086–94. doi: 10.1002/pros.22819. [DOI] [PubMed] [Google Scholar]
- 28.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu XY, Chen LB, Wang JH, Su QS, Yang JR, Lin Y, et al. Overexpression of survivin is correlated with increased invasion and metastasis of colorectal cancer. Journal of surgical oncology. 2012;105:520–8. doi: 10.1002/jso.22134. [DOI] [PubMed] [Google Scholar]
- 30.Tai CJ, Chin-Sheng H, Kuo LJ, Wei PL, Lu HH, Chen HA, et al. Survivin-mediated cancer cell migration through GRP78 and epithelial-mesenchymal transition (EMT) marker expression in Mahlavu cells. Annals of surgical oncology. 2012;19:336–43. doi: 10.1245/s10434-011-1692-5. [DOI] [PubMed] [Google Scholar]
- 31.Knight BB, Oprea-Ilies GM, Nagalingam A, Yang L, Cohen C, Saxena NK, et al. Survivin upregulation, dependent on leptin-EGFR-Notch1 axis, is essential for leptin-induced migration of breast carcinoma cells. Endocrine-related cancer. 2011;18:413–28. doi: 10.1530/ERC-11-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodge DR, Hurt EM, Farrar WL. The role of IL-6 and STAT3 in inflammation and cancer. Eur J Cancer. 2005;41:2502–12. doi: 10.1016/j.ejca.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 33.Grivennikov S, Karin M. Autocrine IL-6 signaling: a key event in tumorigenesis? Cancer Cell. 2008;13:7–9. doi: 10.1016/j.ccr.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 34.Chen M, Wang J, Lu J, Bond MC, Ren XR, Lyerly HK, et al. The anti-helminthic niclosamide inhibits Wnt/Frizzled1 signaling. Biochemistry. 2009;48:10267–74. doi: 10.1021/bi9009677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An J, Wang C, Deng Y, Yu L, Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell reports. 2014;6:657–69. doi: 10.1016/j.celrep.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueda T, Bruchovsky N, Sadar MD. Activation of the androgen receptor N-terminal domain by interleukin-6 via MAPK and STAT3 signal transduction pathways. The Journal of biological chemistry. 2002;277:7076–85. doi: 10.1074/jbc.M108255200. [DOI] [PubMed] [Google Scholar]