Abstract
Background
Next-generation sequencing technologies enable the re-sequencing of a large number of genomes and provide an unprecedented opportunity to discover numerous DNA polymorphisms throughout the genome of a species. As the second most abundant form of genetic variation, InDels, with characteristics of co-dominance, multiple alleles and high stability and density and that are easy to genotype, have received an increasing amount attention.
Results
In this work, a total of 2,329,544 InDels were identified in 1767 rice genomes; these InDels were dispersed across all 12 rice chromosomes, with one InDel marker found, on average, every 160.22 bp. There were 162,380 highly polymorphic InDels with a polymorphism information content (PIC) ≥ 0.5, contributing 1.81 % to the unique primer set. Of these highly polymorphic InDels, we also selected InDels with major allele differences (the size difference between the most and second most frequent alleles) ≥ 3 bp or 8 bp for primer design, which provided a more flexible choice for researchers. Finally, we experimentally validated 100 highly polymorphic InDels for accuracy and polymorphism. The PCR results showed that the accuracy of the InDel markers was 95.70 %, while the average PIC value was 0.56, with a range of 0.19 to 0.78; the average allele number was 3.02, with a range of 2 to 5.
Conclusions
Our genome-wide and easily used InDel markers with high polymorphism and density in both cultivated and wild rice will undoubtedly have practical implications in rice marker-assisted breeding and will also meet the need of fine-scale genetic mapping in map-based rice gene cloning.
Electronic supplementary material
The online version of this article (doi:10.1186/s12284-015-0063-4) contains supplementary material, which is available to authorized users.
Keywords: Rice, Insertion and deletion, Polymorphism markers, Next-generation sequence
Background
Rice (Oryza sativa L.), which is grown worldwide and is an important food crop for more than half of the world’s population, is considered to be the monocot model plant for molecular genetic studies. To be grown successfully under a variety of climatic conditions, breeders maintain rice with high genetic diversity. Molecular markers are useful tools for genetic research and breeding, including genotype fingerprinting, genetic diversity analysis, phylogenetic analysis, map-based gene cloning, variety identification and marker-assisted breeding (McCouch et al. 1997; Joshi et al. 2001; Nagaraju et al. 2002; Ni et al. 2002). A large number of molecular markers have been used in rice, such as restriction fragment length polymorphisms (RFLPs), simple sequence repeat (SSR) markers, single nucleotide polymorphisms (SNPs) and insertions-deletions (InDels) (Akagi et al. 1996; Panaud et al. 1996; Temnykh et al. 2001; McCouch et al. 2002; Nasu et al. 2002; Project IRGS 2005; Ren et al. 2005; Chen et al. 2011). SSRs and SNPs are the two most widely used molecular markers in rice. SSR markers are co-dominant, multi-allelic, easy to use and inexpensive; thus, they have been the most widely used markers in rice for a long period of time (Tautz 1989; McCouch et al. 2002). SNP markers occur at a much higher density in the genome and are amenable to high-throughput methods, such as genotyping arrays (Rafalski 2002). It is worth noting that the low density of SSRs is a major obstacle for their application; large-scale use of SNPs is restricted by less information and alleles as well as high technical or equipment demands. However, on average, the cost of high-density SNP genotyping is relatively low.
As the second most common type of polymorphism and the most abundant structural variant (Mullaney et al. 2010), InDel markers provide a higher density than traditional SSR markers and utilize the same experimental procedure as that used for SSR markers. Because InDel markers have more practical value for laboratories without the infrastructure to perform SNP genotyping (Liu et al. 2013) and because they occur more frequently than SSR markers in the rice genome and use the same experimental approaches that are routinely used for SSR markers, they have received an increasing amount of attention. Recently, InDel markers have been successfully used for genetic studies in wheat (Raman et al. 2006), rice (Hayashi et al. 2006; Ji et al. 2010; Liu et al. 2012), citrus (García-Lor et al. 2012), Arabidopsis (Hou et al. 2010; Păcurar et al. 2012) and natural populations of dogs (Väli et al. 2008). Specifically, for rice, 50 InDel markers developed by Shen et al. (2004) have successfully been used to distinguish the Basmati rice variety from other fragrant rice varieties (Steele et al. 2008). In addition, InDel markers have also been used for the classification of rice indica and japonica subspecies and to examine their relationships regarding evolution (Lu et al. 2009; Liu et al. 2012).
Based on first-generation (Sanger) sequencing, the genomic sequences of two inbred rice cultivars, japonica cv. Nipponbare and indica cv. 9311, were released during the past decade (Goff et al. 2002; Yu et al. 2002). The emergence of next-generation sequencing (NGS) technologies has enabled more efficient re-sequencing of a large number of genomes at a significantly lower cost than ever before and has provided the opportunity to survey high-throughput genotyping and large-scale genetic variation (Weigel and Mott 2009). A massive number of InDel polymorphisms between highly homologous genomes in rice have been identified more efficiently and economically than ever before by the NGS technique (Arai-Kichise et al. 2011). An alignment of shotgun-sequenced contigs of 9311 and the draft genome assembly of Nipponbare revealed 24,557 single-base InDel polymorphisms between the two subspecies, resulting in a polymorphism rate of 0.11 InDels/kb across the whole genome (Feltus et al. 2004). Shen et al. (2004) has constructed a genome-wide DNA polymorphism database using the genome of Nipponbare, a cultivar of japonica, and 9311, a cultivar of indica, which contains 479,406 InDels (Shen et al. 2004).
A large amount of InDel markers have been developed in rice in previous studies, but they lack polymorphism information or allele frequency in different populations or subpopulations. In this article, we collected the re-sequencing data of 1767 worldwide rice varieties from publicly available genomic sequence information to develop genome-wide InDel polymorphism markers through a computational strategy called electronic-PCR (e-PCR) (Schuler 1997); in addition, we experimentally validated some InDel polymorphisms. Thus, the genome-wide and easily used InDel markers with polymorphism information content (PIC) ≥ 0.5 and major allele differences (size difference between the most and second most frequent alleles) ≥ 3 bp or 8 bp, which were clustered by the major groups of O. sativa (Japonica and Indica), O. rufipogon and O. nivara, will have significant practical implications in rice breeding.
Results
Identification and Distribution of the Unique Primer
Using the DNA sequences of the Nipponbare reference genome as the template, we designed 18,662,247 pairs of e-PCR primers based on the sliding window technique, which included 7,738,481 pairs in the intergenic region and 10,923,766 pairs in the genic region, accounting for 41.47 % and 58.53 % of the genic region, respectively. Of the total e-PCR primers, 8,995,927 primer pairs were mapped to the unique genomic regions in both Nipponbare and 9311 simultaneously with a proportion of 48.20 %, which included 3,560,733 pairs in the intergenic region and 5,435,194 pairs in the genic region, accounting for 39.58 % and 60.42 %, respectively. Based on data from Table 1, the density of these unique primers in different genomic regions varied and were ranked in descending order as follows: intron, 3′-UTR (3′-untranslated region), 5′-UTR (5′-untranslated region), TSS_up_0.5 kb (0.5 kb upstream of the transcription start site), intergenic, TES_down_0.5 kb (0.5 kb downstream of the transcription end site) and CDS (coding determining sequences).
Table 1.
Distribution of the primers and InDels in various genomic regions
| Genome Region | Total Primera | Unique Primerb | Unique Primer Setc | InDeld | Highly Polymorphic INDELe | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Count | Count | Densityf | Proportion (%)g | Count | Count | Densityf | Proportion (%)h | Count | Densityf | Proportion (%)i | |
| TSS_up_0.5 kb | 1,372,500 | 659,382 | 23.63 | 48.04 | 655,277 | 227,877 | 8.17 | 34.78 | 17,555 | 0.63 | 2.68 |
| 5′-UTR | 424,611 | 225,331 | 26.66 | 53.07 | 223,737 | 72,931 | 8.63 | 32.60 | 6061 | 0.72 | 2.71 |
| 3′-UTR | 562,748 | 435,385 | 27.64 | 77.37 | 435,014 | 104,248 | 6.62 | 23.96 | 6295 | 0.40 | 1.45 |
| CDS | 3,844,531 | 1,545,879 | 17.48 | 40.21 | 1,536,689 | 158,448 | 1.79 | 10.31 | 4640 | 0.05 | 0.30 |
| Intron | 3,571,402 | 2,106,294 | 39.90 | 58.98 | 2,101,227 | 483,308 | 9.16 | 23.00 | 33,985 | 0.64 | 1.62 |
| TES_down_0.5 kb | 1,329,621 | 643,369 | 23.06 | 48.39 | 641,322 | 192,099 | 6.89 | 29.95 | 13,883 | 0.50 | 2.16 |
| Intergenic | 7,738,481 | 3,560,733 | 23.42 | 46.01 | 3,542,446 | 1,130,446 | 7.44 | 31.91 | 82,495 | 0.54 | 2.33 |
| Total | 18,662,247 | 8,995,927 | 24.10 | 48.20 | 8,955,862 | 2,329,724 | 6.24 | 26.01 | 162,380 | 0.44 | 1.81 |
aTotal primers in various genomic regions
bPrimer pairs simultaneously mapped to unique genomic regions in both Nipponbare and 9311
cA set of unique primers with the e-PCR products in twenty or more than twenty rice genomes
dInDels with PIC > 0
eInDels with PIC ≥ 0.5
fDensity was defined as the corresponding primer or InDel number every 1000 bp
gProportion of unique primers in the total primers
hProportion of InDels in the unique primer set
iProportion of highly polymorphic InDels in the unique primer set
In addition, we also compared the distribution of the unique primers on the 12 rice chromosomes, and the results showed that the count of unique primers on each chromosome was generally consistent with the corresponding chromosome length. The smallest distribution was observed on chromosome 10 (496,144), while the largest distribution was on chromosome 1 (1,175,638). The average density of unique primers dispersed along the 12 rice chromosomes varied from the lowest (19.39 primer pairs/kb, or one primer pair every 51.57 bp) on chromosome 11 to the highest (28.75 primer pairs/kb, or one primer pair every 34.79 bp) on chromosome 3, with an average size of 24.10 primer pairs/kb (one primer pair every 41.49 bp) across all 12 chromosomes (Table 2).
Table 2.
Distribution of the unique primers and InDels on rice chromosomes
| Chromosome | Chromosome length (bp) | Unique primerb | InDeld | ||
|---|---|---|---|---|---|
| Count | Densityf | Count | Densityf | ||
| 1 | 43270923 | 1,175,638 | 27.17 | 299,457 | 6.92 |
| 2 | 35937250 | 1,011,186 | 28.14 | 259,989 | 7.23 |
| 3 | 36413819 | 1,046,792 | 28.75 | 262,345 | 7.20 |
| 4 | 35502694 | 777,218 | 21.89 | 196,803 | 5.54 |
| 5 | 29958434 | 728,667 | 24.32 | 190,707 | 6.37 |
| 6 | 31248787 | 754,761 | 24.15 | 200,616 | 6.42 |
| 7 | 29697621 | 733,083 | 24.68 | 191,446 | 6.45 |
| 8 | 28443022 | 604,746 | 21.26 | 158,366 | 5.57 |
| 9 | 23012720 | 555,769 | 24.15 | 146,065 | 6.35 |
| 10 | 23207287 | 496,144 | 21.38 | 129,305 | 5.57 |
| 11 | 29021106 | 562,780 | 19.39 | 150,050 | 5.17 |
| 12 | 27531856 | 549,143 | 19.95 | 144,395 | 5.24 |
| Total | 373245519 | 8,995,927 | 285.24 | 2,329,544 | 74.04 |
| Average | 31103793.25 | 749,660.58 | 24.10 | 194,128.67 | 6.24 |
bPrimer pairs simultaneously mapped to unique genomic regions in both Nipponbare and 9311
dInDels with PIC > 0
fDensity was defined as the corresponding primer or InDel number every 1000 bp
InDel Identification in Rice Varieties
Based on the next-generation sequencing data from 1765 rice varieties, a total of 7,100,998,643 publicly sequenced reads were downloaded from the NCBI (National Center for Biotechnology Information) website, with an average length of 86.56 bp; the sequencing depth ranged from 0.03× (GP133) to 118.42× (Oryza-brachyantha), with an average depth of 1.89× (Additional file 1: Table S1). Using e-PCR to align 8,995,927 unique primer pairs against publicly sequenced reads of rice varieties, the results showed that the average number of e-PCR products was 1,950,638, with a proportion of 21.68 % (Additional file 2: Table S2), which ranged from 10,631 in Bengal (0.12 %) to 8,764,237 in Omachi (97.42 %). A set of 8,955,862 unique primers had e-PCR products in twenty or more than twenty rice genomes, contributing more than 99.55 % to the total unique primers; then, we identified 2,329,544 InDels based on this set of unique primers, contributing 26.01 % to the unique primer set (Table 1).
InDel Distribution in Rice Genomes
The 2,329,544 InDels were almost evenly distributed across all 12 chromosomes, but the distribution of InDels varied from the smallest amount (129,305) on chromosome 10 to the largest amount (299,457) on chromosome 1. The average density was 6.24 InDels/kb, with a range of 5.17 to 7.23 InDels/kb on 12 chromosomes. The lowest density was on chromosome 11, and the highest density was on chromosome 2 (Table 2). Subsequently, we also looked into the details of InDels in various genomic regions and found that InDels were most abundant in the intergenic region, followed, in order, by intron, TSS_up_0.5 kb, TES_down_0.5 kb, CDS, 3′-UTR and 5′-UTR (Table 1). Thus, the proportion of InDels distributed in the intergenic region was 48.52 %, and the remaining 51.48 % were distributed in the genic region.
The number of InDels decreased with increasing PIC value, and most (1,690,818) had a PIC value with a range from 0 to 0.1, contributing more than 70 % to the total InDels (Fig. 1). With respect to the polymorphism rate (the proportion of InDels in the unique primer set), we concluded that InDels in TSS_up_0.5 kb were the most abundant, followed by 5′-UTR, intergenic, TES_down_0.5 kb, 3′-UTR, intron and CDS (Table 1). In particular, CDS had the lowest polymorphic rate (10.31 %), with the highest (82.82 %) PIC values, varying from 0 to 0.1, while TSS_up_0.5 kb had the highest polymorphic rate (34.78 %) and the fewest (70.84 %) PIC values varying from 0 to 0.1.
Fig. 1.
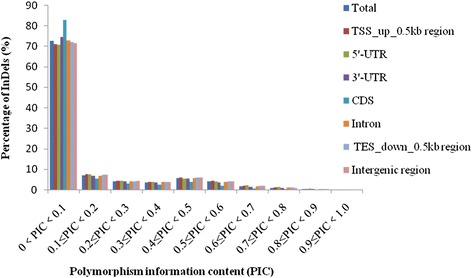
Distribution of InDels in various genomic regions
The number of alleles generated for all InDels varied from 2 to 116, with an average of 2.82. For the InDels distributed in various genomic regions, most (69.66 %) had two alleles, 18.10 % had three alleles, and only a few markers had nine alleles (Fig. 2). Interestingly, for two alleles, CDS had the highest percentage of InDels (83.70 %) and 5′-UTR had the lowest percentage of InDels (63.32 %), but for the remaining allele number, the highest amount of InDels were almost all distributed in the 5′-UTR and the lowest amount were all in the CDS. We also analyzed the polymorphism of the InDel size in various genomic regions and found that the InDel size difference between the shortest and the longest varied from 1 to 214 bp. The majority of the InDel markers (76.50 %) had less than 10-bp size difference: 32.42 % had a size difference of 1 bp, 13.53 % had a size difference of 2 bp, 9.03 % had a size difference of 3 bp, 6.57 % had a size difference of 4 bp, 3.64 % had a size difference of 5 bp, 3.56 % had a size difference of 6 bp, 2.67 % had a size difference of 7 bp, 2.96 % had a size difference of 8 bp and 2.11 % had a size difference of 9 bp. Only 23.50 % of the markers had a size difference of more than 10 bp. Thus, the size differences of 1 and 2 bp made up a higher percentage (45.95 %) than the size differences from 3 bp to 9 bp (30.54 %). For various genomic regions, except the CDS region, all had a declining percentage with increasing InDel size difference (Fig. 3).
Fig. 2.
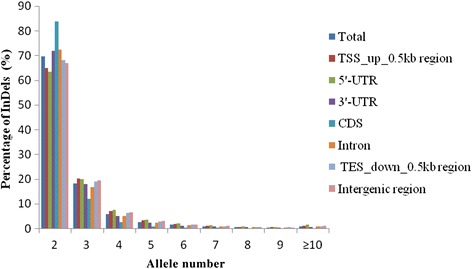
Distribution of allele number in various genomic regions
Fig. 3.
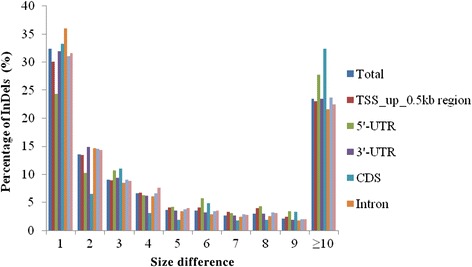
Distribution of InDel size difference in various genomic regions
Primer Design for New InDel Markers
For different resolutions of polyacrylamide gel electrophoresis and agarose gel electrophoresis, two sets of highly polymorphic InDel markers were selected from 162,380 highly polymorphic InDels with PIC ≥ 0.5. One set of 41855 InDels with major allele difference ≥ 3 bp and PCR product lengths of 60–100 bp was selected for polyacrylamide gel electrophoresis (Additional file 3: Table S3), while the other set of 14428 InDels with a major allele difference ≥ 8 bp and PCR product lengths of 150–300 bp was selected for agarose gel electrophoresis (Additional file 4: Table S4). The exact positions of these InDels, as well as information on the PCR-based primers and PIC are presented in Table S3 and Table S4. These positions would be very useful for primer selection in actual practice, and researchers could also design primers from the tag sequences surrounding the InDels. In addition, major allele frequencies within the major groups of O. sativa (Japonica and Indica), O. rufipogon and O. nivara and the gene ID and annotation, if available, are included in Table S3 and Table S4 as well. Finally, we also provided an ontological analysis of the genes affected by I with PIC ≥ 0.5 and a major allele difference ≥3 bp in the CDS region (Additional file 5: Table S5); these characteristics made these InDels valuable for molecular breeding applications.
Experimental Validation
A set of 100 InDel markers that were evenly spread across the rice genome were selected, and primers were designed for experimental validation (Additional file 6: Table S6). The 100 InDel markers were developed by e-PCR based on the InDel polymorphisms between the Nipponbare and 9311 reference sequences; thus, these markers should theoretically be polymorphic between Nipponbare and 9311. To test their accuracy, we amplified the genomic DNA of Nipponbare and 9311 by PCR. The PCR results showed that only seven primer pairs could not simultaneously generate PCR products from the genomic DNA of both Nipponbare and 9311, and 89 primer pairs were polymorphic between Nipponbare and 9311; thus, the PCR success rate was 93 %, while the accuracy of InDel markers was 95.70 %.
To test their polymorphisms in other rice varieties, we further analyzed them by genomic DNA PCR amplification in a panel of 20 rice cultivars, including Nipponbare and 9311, using the same set of InDel markers plus 100 pairs of SSR primers for comparison (Additional file 7: Table S7; Fig. 4). For the InDel markers, their average accuracy was approximately 98.2 % in 20 rice cultivars, while the average PIC value was 0.56, with a range of 0.19 to 0.78, and the average allele number was 3.02, with a range of 2 to 5, using the genomic DNA of 20 rice cultivars as the template for PCR amplification. However, the PIC value varied from 0.50 to 0.95, with an average of 0.78, and the allele number varied from 2 to 34, with an average of 12.25, using the re-sequencing data of 1765 rice varieties as the template for e-PCR. For 100 pairs of SSR primers, the PIC value ranged from 0 to 0.78, with an average of 0.35, and the allele number ranged from 1 to 5, with an average of 2.17 (Additional file 8: Table S8).
Fig. 4.

Experimental validation for two InDels on 4-33854060 and 5-1279380. PCR product from lane1 to lane20 was Nipponbare, 9311, zaomadao, SRSye, beixiang7, 60kang, 72gan, 955R, T116, sanpang76, R1318, nanhui511, guiyangai49, qingnongai, huangxinzhan, yanjing2, yandao8, xudao3, huaidao13 and wuyunjing3, respectively. (The number below horizontal line showed chromosome and physical position. M: Marker DL2000)
Discussion
The emergence of next-generation sequencing technologies has enabled more efficient genome sequencing at a significantly lower cost than ever before and has provided the community with the opportunity to find genome-wide DNA polymorphisms for genetic studies. In this article, we examined 1767 rice genomes, representing a much wider sample than has previously been used, and detected a high number of InDels across the rice genome, which was nearly three- and four-fold more than in previous work (Xu et al. 2012; Huang et al. 2012); thus, we developed the most broadly representative high-density InDel markers, with one InDel every 160 bp along the rice genome, which was nearly 65-fold of that of genes (Project IRGS 2005), one-thousand times denser than SSR and six times denser than other InDels developed in previous studies (Shen et al. 2004). Accordingly, these genome-wide InDel polymorphism markers could meet the need of fine-scale genetic mapping in map-based cloning of rice genes. Notably, more than half of the InDels are distributed in genic regions, and these InDels are considered to be genic molecular markers or functional markers (Varshney et al. 2007) that are superior to random DNA markers, such as RFLPs, SSRs and AFLPs, because of complete linkage with trait locus alleles (Andersen and Lübberstedt 2003). In addition, genic molecular markers have provided opportunities to examine functional diversity, and with the development of more genic molecular markers in major crop species, genetic diversity studies will become more meaningful.
Except for high density, high polymorphism is the most important evaluation factor for marker development. Our genome-wide InDel polymorphism markers also contains highly polymorphic InDel markers (PIC ≥ 0.5), and PCR validation has proved our markers to be highly polymorphic. The average polymorphism and allele number of the InDels are both higher than those of SSR in PCR validation, but lower than those in e-PCR. This is mainly because the gel resolution is lower than e-PCR and because e-PCR can detect even one base difference. In addition, the detection efficiency of gel electrophoresis not only requires a size difference but also demands an amplicon length. The detection efficiency is enhanced by increasing the ratio of the size difference to amplicon length. In this study, we have developed two sets of highly polymorphic InDel markers for polyacrylamide gel electrophoresis and agarose gel electrophoresis so that researchers can determine which InDels may be useful, depending on their specific programs, by downloading our data without any limitations. In summary, the InDels with a high amount of polymorphisms and a large size difference will be useful as density markers of genome variation for marker-assisted mapping of important rice traits as well as for rice breeding, and the data generated in this study will be a valuable resource for rice improvement.
Moreover, the precise identification of InDels in sequence databases depends on the strategy and parameters used for data mining, thus these two points might be noteworthy. First, we adopted a computational strategy called e-PCR in this study; e-PCR can not only virtually simulate the PCR process but can also save time and reduce laboratory costs using in silico assays. Specifically, when developing new markers for mapping studies, e-PCR can be used to test potential primers in various ways before actually incurring the expense of oligonucleotide synthesis. Compared to variant identification software, such as samtools and GATK, which directly reads the map of the reference sequence, e-PCR utilizes hash-based string searching instead of whole sequence alignment. Second, the most important parameter for unique primer identification in e-PCR is fault tolerance. In this study, fault tolerance is set to three in e-PCR pipeline Step 2 to remove multiple loci as much as possible and increase the probability of the unique primers being aligned to reads from samples, while fault tolerance is set to one in e-PCR pipeline Step 3, given the tradeoff between InDel accuracy and SNP tolerance. Accordingly, the methods mentioned above greatly reduced the potential false positives in this article.
Finally, a much broader sample data set that was collected from publicly available genomic sequence information will also provide an opportunity to develop millions of InDels in representative wild and cultivated rice. The cultivated rice are broadly representative varieties with yield-related traits, quality-related traits and resistance-related traits, while the wild species of the genus Oryza contain a largely untapped reservoir of agronomically important genes for rice improvement. Thus, we developed genome-wide InDel polymorphism markers in both cultivated and wild rice that would greatly facilitate marker-assisted breeding and rice gene mapping.
Conclusion
In this study, we developed a large amount of genome-wide and easily used InDel markers with a high amount of polymorphisms and density. The identification and validation of these InDel polymorphism markers among 1767 rice genomes will provide molecular markers for genetic study, such as gene cloning and association analysis, as well as marker-assisted selection in breeding.
Methods
Data Sources of Rice Genomic Sequences
We downloaded the genomic sequence data of two rice cultivars, Nipponbare (MSU version Release 7) and 9311, from the websites http://rice.plantbiology.msu.edu/index.shtml and http://rice.genomics.org.cn/rice/link/download.jsp. The 5′-UTR, CDS, 3′-UTR, exon, intron and intergenic sequences were provided by the annotation of the Nipponbare genome (http://rice.plantbiology.msu.edu/index.shtml). In addition, we also downloaded the re-sequencing data of 1765 rice varieties from the NCBI web site http://www.ncbi.nlm.nih.gov/sra?term=ERA000213, SRA023116, ERA081008, SRP026477, ERP000729, SRA051809, SRA036041, SRA036085, DRR000719, ERA009071, and SRA046411. All 1767 rice genotypes were composed of cultivated rice (Oryza sativa L.) and its wild relatives; thus, these inbred lines were selected to represent a diverse collection (Additional file 1: Table S1).
Plant Materials and DNA Isolation
A panel of 20 rice cultivars, including 6 japonica-type rice cultivar (Nipponbare, yanjing2, yandao8, xudao3, huaidao13, wuyunjing3) and 14 indica-type rice cultivar (9311, zaomadao, SRSye, beixiang7, 60kang, 72gan, 955R, T116, sanpang76, R1318, nanhui511, guiyangai49, qingnongai, huangxinzhan), were used to validate InDel polymorphisms. Genomic DNA was isolated from 2-week-old seedlings using the modified CTAB (Cetyltrimethyl Ammonium Bromide) method (Murray and Thompson 1980).
e-PCR Primer Design and Unique Primer Identification
Using the DNA sequences of the Nipponbare reference genome as the template, we designed e-PCR primers by a Perl script based on the sliding window technique. In the sliding window technique, a defined window is slid using each previous primer pair at a time along the template sequence, and then, a set of primer pairs are designed to cover the entire genome as much as possible (Yang et al. 2011). First, we extracted 20 bp from the first nucleotide of the template sequence as the forward primer, and after an interval of 20 bp, we extracted another 20 bp sequence as the reverse primer for the designed primer pair within the sliding window. Subsequently, the sliding window shifted based on the previous reverse primer, and the next forward primer was designed from the 20-bp upstream region of the previous reverse primer (Fig. 5). Thus, the remaining primers were designed in the same way, and this procedure continued until the entire template sequence was covered by all primer pair sets (Fig. 6 Step 1). After the sliding window process was performed, the sequences of primer pairs were aligned to the Nipponbare and 9311 reference genomes with Bowtie allowing up to three mismatches (Fig. 6 Step 2). Unique primers in both reference genomes simultaneously were also identified by a Perl script (Langmead et al. 2009).
Fig. 5.
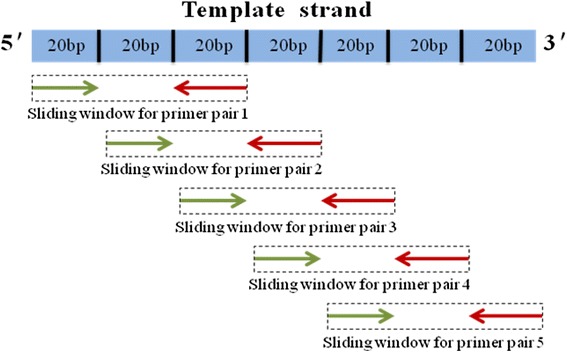
e-PCR primer design based on sliding window. The green arrow lines were the forward primers and the red arrow lines were the reverse primers
Fig. 6.
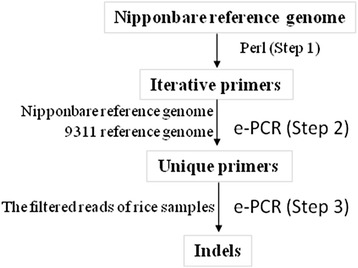
e-PCR Pipeline
InDel Identification in Rice Varieties
We used NGSQC toolkit v2.3.3 to filter for high quality data (cutoff quality score of 20) (Patel and Jain 2012). Using the re-sequencing reads from 1765 rice varieties as the template, unique primer sequences were aligned with Bowtie, allowing up to one mismatch (Fig. 6 Step 3). If there were many e-PCR amplicon products for the same allelic locus, the most frequent one was selected by a Perl script, which was similar to the strategy for contig assembling in ABySS. The allelic diversity of each InDel with the e-PCR products in twenty or more than twenty rice genomes was assessed by PIC, which was defined as , where pij is the frequency of the jth pattern for the ith marker (Anderson et al. 1993).
Analysis of Enriched Gene Ontologies
We used AgriGO (Du et al. 2010) at the default setting to examine the significant enrichment GO terms for each gene affected by InDels with PIC ≥ 0.5 and major allele differences ≥3 bp in the CDS region, and the gene annotation used in this study was obtained from MSU version Release 7.
PCR Primer Design
To design primers for PCR validation, sequences with a total of 100 bp, including a 20-bp InDel region and flanking sequences of 40 bp for each side of the InDel region, were extracted. Primers were designed using Primer3 (Rozen and Skaletsky 2000), with a primer length of 20 nt to 28 nt and an optimal length of 23 nt, melting temperature (Tm) of 60 to 65 °C and an optimal temperature of 63 °C, PCR product length of 60 to 100 bp and ending with G- or C-rich at the 3′-end. Several primer pairs were designed for each InDel. The best primer pair was selected based on a similar Tm value with a GC content of 30-70 % and an optimal GC content of 50 %. All of the processes were carried out using custom Perl scripts.
Experimental Validation of InDels
A set of 100 InDel markers (Additional file 6: Table S6) with PIC ≥ 0.5 and a 3–10 bp major allele difference in genomes of two inbred rice cultivars between Nipponbare and 9311 were randomly selected from the developed InDel markers for accuracy and polymorphism validation by the PCR technique. All 100 InDel markers were first used to amplify the genomic DNA of Nipponbare and 9311 for InDel accuracy validation. Then, we further detected their polymorphisms by PCR amplification of genomic DNA in a panel of 20 rice cultivars, including Nipponbare and 9311. For comparison purposes, 100 pairs of SSR primers were randomly selected (Additional file 7: Table S7), and the PIC value for each SSR marker was calculated using the formula described above.
PCR was performed in a 15-μL reaction volume containing 50 ng of template DNA, 1.5 μL of 10 × buffer (Mg2+), 2.0 μL of dNTP (2.5 mM), 100 nM of each SSR-primer, 2 U of Taq polymerase, and ddH2O. The DNA amplification protocol included an initial denaturation for 3 min at 95 °C, followed by 35 cycles of denaturation for 30 s at 95 °C, annealing for 90 s at 55 °C, and an extension for 90 s at 72 °C, with a final extension for 10 min at 72 °C. The reactions were performed in a C1000 thermal cycler (Bio-rad, Inc., Hercules, CA). The PCR products were subsequently separated in 6.0 % polyacrylamide gel and visualized using the silver-staining approach (Bassam et al. 1991).
Acknowledgements
We thank Shuangcheng Li from Rice Research Institute of Sichuan Agricultural University for providing DNA from a panel of 20 rice cultivars used in this study and technical assistance.
Abbreviations
- RFLP
Fragment length polymorphism
- SSR
Simple sequence repeat
- SNP
Single nucleotide polymorphism
- InDel
Insertions-deletion
- NGS
Next-generation sequencing
- e-PCR
Electronic PCR
- 3′-UTR
3′-untranslated region
- 5′-UTR
5′-untranslated region
- TSS_up_0.5 kb
0.5 kb upstream of the transcription start site
- TES_down_0.5 kb
0.5 kb downstream of the transcription end site
- CDS
Coding determining sequences
- NCBI
National Center for Biotechnology Information
- PIC
Polymorphism information content
- Major allele difference
Size difference between the most and second most frequent alleles
- InDel size difference
Size difference between the shortest and the longest InDel
- Polymorphism rate
The proportion of InDels in the unique primer set
- CTAB
Cetyltrimethyl Ammonium Bromide
Additional files
Description of rice genotypes for e-PCR. (XLS 357 kb)
e-PCR products of rice genotypes. (XLS 208 kb)
InDels with major allele difference ≥3 bp. (XLS 19671 kb)
InDels with major allele difference ≥8 bp. (XLS 10074 kb)
Ontology analysis of genes affected by InDels with PIC ≥ 0.5 and major allele difference ≥3 bp in CDS region. (XLS 85 kb)
Chromosomal position and primer sequence for 100 InDel markers. (XLS 37 kb)
Chromosomal position and primer sequence for 100 SSRs. (XLS 33 kb)
Allele number and polymorphic information content for InDel and SSR. (XLS 43 kb)
Footnotes
Jian Liu Jingwei Li and Jingtao Qu contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JL conceived of the study and corrected the manuscript. JWL designed and wrote the paper. JTQ performed bioinformatics analysis. SYY participated in validation. All of the authors read and approved the final manuscript.
Contributor Information
Jian Liu, Phone: +86-28-86290916, Email: liujian@sicau.edu.cn.
Jingwei Li, Email: llijingwei@163.com.
Jingtao Qu, Email: 757071279@qq.com.
Shuangyong Yan, Email: yzxm2000@yahoo.com.cn.
References
- Akagi H, Yokozeki Y, Inagaki A, Fujimura T. Microsatellite DNA markers for rice chromosomes. Theor Appl Genet. 1996;93(7):1071–1077. doi: 10.1007/BF00230127. [DOI] [PubMed] [Google Scholar]
- Andersen JR, Lübberstedt T. Functional markers in plants. Trends Plant Sci. 2003;8(11):554–560. doi: 10.1016/j.tplants.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Anderson JA, Churchill G, Autrique J, Tanksley S, Sorrells M. Optimizing parental selection for genetic linkage maps. Genome. 1993;36(1):181–186. doi: 10.1139/g93-024. [DOI] [PubMed] [Google Scholar]
- Arai-Kichise Y, Shiwa Y, Nagasaki H, Ebana K, Yoshikawa H, Yano M, Wakasa K. Discovery of genome-wide DNA polymorphisms in a landrace cultivar of japonica rice by whole-genome sequencing. Plant Cell Physiol. 2011;52(2):274–282. doi: 10.1093/pcp/pcr003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassam BJ, Caetano-Anollés G, Gresshoff PM. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196(1):80–83. doi: 10.1016/0003-2697(91)90120-I. [DOI] [PubMed] [Google Scholar]
- Chen H, He H, Zou Y, Chen W, Yu R, Liu X, Yang Y, Gao Y-M, Xu J-L, Fan L-M. Development and application of a set of breeder-friendly SNP markers for genetic analyses and molecular breeding of rice (Oryza sativa L.) Theor Appl Genet. 2011;123(6):869–879. doi: 10.1007/s00122-011-1633-5. [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. AgriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltus FA, Wan J, Schulze SR, Estill JC, Jiang N, Paterson AH. An SNP resource for rice genetics and breeding based on subspecies indica and japonica genome alignments. Genome Res. 2004;14(9):1812–1819. doi: 10.1101/gr.2479404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Lor A, Luro F, Navarro L, Ollitrault P. Comparative use of InDel and SSR markers in deciphering the interspecific structure of cultivated citrus genetic diversity: a perspective for genetic association studies. Mol Gen Genomics. 2012;287(1):77–94. doi: 10.1007/s00438-011-0658-4. [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan T-H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296(5565):92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshida H, Ashikawa I. Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes. Theor Appl Genet. 2006;113(2):251–260. doi: 10.1007/s00122-006-0290-6. [DOI] [PubMed] [Google Scholar]
- Hou X, Li L, Peng Z, Wei B, Tang S, Ding M, Liu J, Zhang F, Zhao Y, Gu H. A platform of high‐density INDEL/CAPS markers for map‐based cloning in Arabidopsis. The Plant Journal. 2010;63(5):880–888. doi: 10.1111/j.1365-313X.2010.04277.x. [DOI] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, Wang Z-X, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490(7421):497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Lu J, Chao Q, Zhang Y, Zhang M, Gu M, Xu M. Two sequence alterations, a 136 bp InDel and an A/C polymorphic site, in the S5 locus are associated with spikelet fertility of indica-japonica hybrid in rice. Journal of Genetics and Genomics. 2010;37(1):57–68. doi: 10.1016/S1673-8527(09)60025-4. [DOI] [PubMed] [Google Scholar]
- Joshi S, Bhave S, Chowdari K, Apte G, Dhonukshe B, Lalitha K, Ranjekar P, Gupta V. Use of DNA markers in prediction of hybrid performance and heterosis for a three-line hybrid system in rice. Biochem Genet. 2001;39(5–6):179–200. doi: 10.1023/A:1010293325482. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Cai XX, Lu BR. Single‐seeded InDel fingerprints in rice: An effective tool for indica–japonica rice classification and evolutionary studies. J Syst Evol. 2012;50(1):1–11. doi: 10.1111/j.1759-6831.2011.00173.x. [DOI] [Google Scholar]
- Liu B, Wang Y, Zhai W, Deng J, Wang H, Cui Y, Cheng F, Wang X, Wu J. Development of InDel markers for Brassica rapa based on whole-genome re-sequencing. Theor Appl Genet. 2013;126(1):231–239. doi: 10.1007/s00122-012-1976-6. [DOI] [PubMed] [Google Scholar]
- Lu B-R, Cai X, Xin J. Efficient indica and japonica rice identification based on the InDel molecular method: Its implication in rice breeding and evolutionary research. Prog Nat Sci. 2009;19(10):1241–1252. doi: 10.1016/j.pnsc.2009.01.011. [DOI] [Google Scholar]
- McCouch SR, Chen X, Panaud O, Temnykh S, Xu Y, Cho YG, Huang N, Ishii T, Blair M. Microsatellite marker development, mapping and applications in rice genetics and breeding. Plant Mol Biol. 1997;35(1–2):89–99. doi: 10.1023/A:1005711431474. [DOI] [PubMed] [Google Scholar]
- McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.) DNA Res. 2002;9(6):199–207. doi: 10.1093/dnares/9.6.199. [DOI] [PubMed] [Google Scholar]
- Mullaney JM, Mills RE, Pittard WS, Devine SE. Small insertions and deletions (INDELs) in human genomes. Hum Mol Genet. 2010;19(R2):R131–R136. doi: 10.1093/hmg/ddq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8(19):4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraju J, Kathirvel M, Kumar RR, Siddiq E, Hasnain SE. Genetic analysis of traditional and evolved Basmati and non-Basmati rice varieties by using fluorescence-based ISSR-PCR and SSR markers. Proc Natl Acad Sci. 2002;99(9):5836–5841. doi: 10.1073/pnas.042099099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu S, Suzuki J, Ohta R, Hasegawa K, Yui R, Kitazawa N, Monna L, Minobe Y. Search for and analysis of single nucleotide polymorphisms (SNPs) in rice (Oryza sativa, Oryza rufipogon) and establishment of SNP markers. DNA Res. 2002;9(5):163–171. doi: 10.1093/dnares/9.5.163. [DOI] [PubMed] [Google Scholar]
- Ni J, Colowit PM, Mackill DJ. Evaluation of genetic diversity in rice subspecies using microsatellite markers. Crop Sci. 2002;42(2):601–607. doi: 10.2135/cropsci2002.0601. [DOI] [Google Scholar]
- Păcurar DI, Păcurar ML, Street N, Bussell JD, Pop TI, Gutierrez L, Bellini C. A collection of INDEL markers for map-based cloning in seven Arabidopsis accessions. J Exp Bot. 2012;63(7):2491–2501. doi: 10.1093/jxb/err422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaud O, Chen X, McCouch S. Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.) Mol Gen Genet MGG. 1996;252(5):597–607. doi: 10.1007/BF02172406. [DOI] [PubMed] [Google Scholar]
- Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7(2):e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Project IRGS The map-based sequence of the rice genome. Nature. 2005;436(7052):793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Rafalski A. Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol. 2002;5(2):94–100. doi: 10.1016/S1369-5266(02)00240-6. [DOI] [PubMed] [Google Scholar]
- Raman H, Raman R, Wood R, Martin P. Repetitive indel markers within the ALMT1 gene conditioning aluminium tolerance in wheat (Triticum aestivum L.) Mol Breed. 2006;18(2):171–183. doi: 10.1007/s11032-006-9025-2. [DOI] [Google Scholar]
- Ren Z-H, Gao J-P, Li L-G, Cai X-L, Huang W, Chao D-Y, Zhu M-Z, Wang Z-Y, Luan S, Lin H-X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37(10):1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. New Jersey: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Schuler GD. Sequence mapping by electronic PCR. Genome Res. 1997;7(5):541–550. doi: 10.1101/gr.7.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y-J, Jiang H, Jin J-P, Zhang Z-B, Xi B, He Y-Y, Wang G, Wang C, Qian L, Li X. Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol. 2004;135(3):1198–1205. doi: 10.1104/pp.103.038463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele KA, Ogden R, McEwing R, Briggs H, Gorham J. InDel markers distinguish Basmatis from other fragrant rice varieties. Field Crop Res. 2008;105(1):81–87. doi: 10.1016/j.fcr.2007.08.001. [DOI] [Google Scholar]
- Tautz D. Hypervariabflity of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989;17(16):6463–6471. doi: 10.1093/nar/17.16.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11(8):1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Väli Ü, Brandström M, Johansson M, Ellegren H. Insertion-deletion polymorphisms (indels) as genetic markers in natural populations. BMC Genet. 2008;9(1):8. doi: 10.1186/1471-2156-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Mahendar T, Aggarwal RK, Börner A (2007) Genic molecular markers in plants: development and applications. In: Genomics-assisted crop improvement. Springer, pp 13–29
- Weigel D, Mott R. The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 2009;10(5):107. doi: 10.1186/gb-2009-10-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X, Dong Y, Gutenkunst RN, Fang L, Huang L. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol. 2012;30(1):105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- Yang C-H, Chang H-W, Ho C-H, Chou Y-C, Chuang L-Y. Conserved PCR primer set designing for closely-related species to complete mitochondrial genome sequencing using a sliding window-based PSO algorithm. PLoS One. 2011;6(3):e17729. doi: 10.1371/journal.pone.0017729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK-S, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296(5565):79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]


