Abstract
Although IL-18 has not previously been shown to promote T lymphopoiesis, results obtained via a novel data mining algorithm (GAMMA), led us to explore a predicted role for this cytokine in T cell development. IL-18 is a member of the IL-1 cytokine family that has been extensively characterized as a mediator of inflammatory immune responses. To assess a potential role for IL-18 in T cell development, we sort-purified mouse bone marrow derived common lymphoid progenitor cells (CLP), early thymic progenitors (ETP) and DN2 thymocytes and cultured these populations on OP9-DL4 stromal layers in the presence or absence of IL-18 and/or IL-7. After one week of culture, IL-18 promoted proliferation and accelerated differentiation of ETPs to the DN3 stage, similar in efficiency to IL-7. IL-18 showed synergy with IL-7 and enhanced proliferation of both the thymus derived progenitor cells and the bone marrow derived common lymphoid progenitor cells. The synergistic effect on the ETP population was further characterized and found to correlate with increased surface expression of c-Kit and IL-7 receptors on the IL-18-treated cells. In summary, we successfully validated the GAMMA prediction that IL-18 affects T lymphopoiesis and demonstrated that IL-18 can positively impact bone marrow lymphopoiesis and T cell development, presumably via interaction with the c-Kit and IL-7 signaling axis.
Introduction
Using a bioinformatics approach (1), IL-18 was predicted to be involved in T cell development and differentiation by a program called GAMMA (Global Microarray Meta-Analysis). GAMMA performs a meta-analysis of ~16,600 publicly available microarray experiments, across different technological platforms (2-color, 1-color, and RNAseq), to identify and rank gene pairs that are strongly correlated across experimental conditions (1). The approach identifies sets of genes that are frequently transcribed and repressed together, regardless of the experimental condition being analyzed, implying the pair is required for the same biological purpose, whether such a relationship is formally documented or not. To identify what these sets of genes have in common, an automated literature-mining program, IRIDESCENT (Implicit Relationship IDEntification by in-Silico Construction of an Entity based Network from Text) (2–4), was used to find published associations the gene set has with diseases, phenotypes and function. GAMMA performed well on computational benchmarks using genes of known function and was also successfully used to predict function for several different poorly characterized or uncharacterized genes that were subsequently validated by laboratory experiments(5–9). Of the genes that were identified using this predictive strategy when employed for discovery of novel factors regulating thymopoiesis, IL-18 was of interest because it has not been directly linked to early T cell development.
IL-18 was originally described as IFN-γ-inducing factor because it was able to augment the production of IFN-γ from T cells and NK cells (10). As a part of the IL-1 cytokine family, IL-18 is a multi-functional component of both the innate and the acquired immune response. Under various conditions the IL-18R1 and IL-18RAP (IL-18Receptor Accessory Protein) are expressed on a variety of immune cells including NK cells, macrophages, neutrophils, B cells, and fully differentiated Th1 cells [reviewed in (11)]. IL-18 has been shown to work in synergy with other cytokines, including IL-12 and IL-4 and has been broadly implicated in autoimmune and inflammatory diseases as well as chronic allergic rhinitis and asthma (12). In the periphery, IL-18 is known to exert an influence on numerous and diverse T cell processes. It increases Fas ligand-mediated cytotoxicity on T cells (13) and stimulates the development of CD8 effector T cells (14). IL-18 also promotes chemotaxis of T cells (15). Furthermore, IL-18 drives CD4 T cell effector responses; inducing IFN-γ production by Th1 cells and promoting production of IL-4, IL-5 and IL-13 in Th2 cells (16–18). IL-18 can also enhance Th2 responses (with IL-2) and is indispensable for Th17 responses (19). Transgenic overexpression of IL-18 had dramatic effects on the immune system, however, these studies did not focus on the effects on early thymocytes, perhaps due to the important role for this cytokine in Th1 and Th2 differentiation that has kept the spotlight on peripheral immune cell mechanisms (20, 21). Although the immunomodulatory functions of IL-18 are relatively well defined, its potential role in T cell development, as predicted by GAMMA is not known. Previous studies have demonstrated thymic expression of IL-18 and this cytokine has been shown to promote the differentiation of fetal DN thymocytes to thymic-derived dendritic cells (22, 23). Furthermore, thymocyte stimulation with IL-18 can elicit production of Th1 and Th2 cytokines in the presence of IL-12 and IL-2, respectively (24). These studies demonstrated the potential of IL-18 to signal within the thymic microenvironment and indicated that IL-18 may indeed be an as yet unrecognized factor capable of influencing early T cell development.
In an attempt to assess the potential role of IL-18 in T cell development, we co-cultured mouse bone marrow HSC and CLP and thymus ETP and DN2 on OP9-DL4 stromal cells, either with IL-18 alone or in conjunction with IL-7, which is traditionally used to promote proliferation and survival of thymocytes in this system (25). We found that IL-18 synergized with IL-7 in promoting strong proliferation of CLP, ETP and DN2 populations. The effect on the ETP cells was further characterized and found to correlate with increased surface expression of CD127 and CD117 on these cells. Surprisingly, we found that IL-18 alone was capable of promoting ETP proliferation to a magnitude similar to that observed for IL-7. These findings demonstrate a novel role for IL-18 in promoting in vitro T cell development from immature precursors and further validate GAMMA as a method to predict putative phenotype and function for genes.
Materials and Methods
Mice
C57BL/6 were bred and housed at the University of Oklahoma-Tulsa Comparative Medicine satellite facility under the oversight of the University of Oklahoma Health Science Center Comparative Medicine Facility (Oklahoma City, OK), an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) approved animal facility. Animal husbandry and all experiments were performed in accordance with procedures outlined in the Guide for the Care and Use of Laboratory Animals (National Research Council). Protocols were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Oklahoma Health Science Center. Mice used in this study were females ranging from 6 to 12 weeks of age. IL18r1 deficient mice (Strain B6.129P2-Il18r1tm1AKI/J) on a C57BL/6 background (26) were purchased from the Jackson Laboratory (Bar Harbor, Maine).
Tissue harvest and cell staining
Thymuses were harvested and placed into complete tumor media (CTM) as previously described (27). Thymuses were crushed through 70-μm nylon cell strainers to produce single thymocyte suspensions. Cells were treated with RBC lysis buffer (Sigma-Aldrich, St. Louis, MO) and washed into CTM prior to counting. Thymocytes at a concentration of 1×108 cells/ml, were incubated with mAb against mouse CD16/CD32 (Fc Block) (BD Biosciences, San Jose, CA) to block potential Fc-mediated binding and then stained at a density of 1×108 cell/ml with primary mAbs for DN3a, DN3b and DN4a sorts: CD4-bio, CD8-bio, TCRγδ-bio, TCR-β-bio, Lin-bio (28), CD25-PE, CD44-APC-Cy7, CD28-FITC for 45 minutes at 4°C in the dark. After two washes, the cells were further stained with SA-PE Texas Red for 30 minutes at 4°C in the dark. ISP (CD4−CD8+CD24hiTCR-β−) cells were sorted-purified by staining with CD4-APC, CD8-FITC, TCR-β-PE-Cy7, and CD24-PE. ETP and DN2 cells were sort-purified, as shown in figure 1A, by staining with the following fluorochrome and biotin coupled mAbs followed by SA-PE Texas Red: CD4-bio, CD8-bio, TCRγδ-bio, TCR-β-bio Lin-bio (28), CD25-PE, CD44-APC-Cy7, c-kit-FITC. To assess the proliferation kinetics, ETP were labeled with CFSE (Life Technologies, Grand Island, NY) prior to placing in co-culture. CFSE labeling was performed by incubating cells at room temperature for 10 min in PBS solution containing 1% FBS and 5μM CFDA-SE and removing excess CFDA-SE by washing cells with culture media. 1,000 cells from each population were cultured in replicate wells in a 24-well plate containing confluent OP9-DL4 stromal cells with/without cytokine(s). All cytokines were obtained from R&D Systems (Minneapolis, MN). Unless otherwise indicated cytokine concentrations were as follows: IL-7 (5 ng/ml), and IL-18 (100 ng/ml).
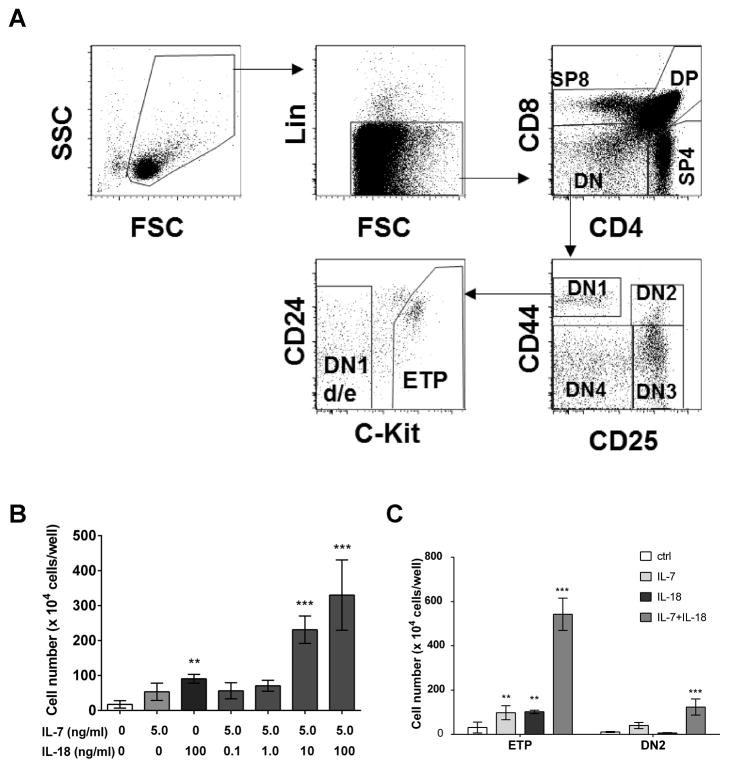
Figure 1. Enhanced ETP and DN2 thymocyte expansion in the presence of IL-18.
A) Gating strategy used for discriminating ETP and DN2 thymocyte subsets. Sort-purified ETP or DN2 thymocytes were co-cultured on OP9-DL4 stromal cells in culture media supplemented with IL-18 or IL-7 alone or in combination and cell yields on day 7 were measured using a hemocytometer. B) Cell number in ETP cultures stimulated with indicated concentrations of IL-7 and IL-18. C) Cell yields in ETP and DN2 cultures stimulated with 5ng/ml IL-7 and 100ng/ml IL-18. Data are presented as mean ± SEM of 3 replicate wells for each experimental condition and are representative of three experiments with similar results. ***p <0.001, **p <0.05.
For hematopoietic stem cells (HSC) (Lin−Kit+CD127−) and common lymphoid progenitor cells (CLP) (Lin−Kit+CD127+) isolation, single cell suspensions of bone marrow from WT mice were processed to remove RBCs and block potential FC-mediated antibody binding by treating cells with RBC lysis buffer followed by staining with anti-CD16/CD32 antibody. Bone marrow cells (1 × 108 cells/ml) were then stained with a biotin-conjugated lineage cocktail (CD45RA (clone:14.8), Gr1 (clone:RB6-8C5), CD11b (clone: M1/70), Ter119 (clone: TER-119), CD45/B220 (clone: RA3-6B2), CD2 (clone RM2-5), CD3 (clone: 145-2C11), Cd8 (clone: 53.6.7), CD49b (clone: DX5), and CD19 (clone:1D3)) and anti-mouse mAb against c-kit (c-kit-APC) and IL-7Ra (CD127-PE). This staining was followed by incubation with SA-PE Texas Red. Based on these markers, HSC and CLP were discriminated and sort-purified.
After times indicated in the Results section, the co-cultured cells were harvested by aspirating and discarding half the culture volume then re-suspending the co-cultured cells by forceful pipetting in the remaining culture volume followed by straining through a 30 μm mesh to remove any detached OP9-DL4 monolayer cell aggregates from the plate. Any remaining OP9-DL4 cells were further discriminated from thymocytes by gating using forward and side scatter; gating out the much larger OP9 cells. The cells were stained with CD4-APC, CD8-Pacific Blue, CD25-PE, CD44-APC-Cy7, CD28-FITC, TCR-β-PE-Cy7, TCRγδ-bio, Lin-bio (28), followed by SA-PE Texas Red. To assess changes in IL-18R1, CD117, and CD127 surface expression, cells were stained with FITC-conjugated anti-IL-18R1 (R&D system; clone 112614), APC conjugated anti-CD117 (Biolegend; clone 2B8), or PE conjugated anti-CD127 (eBioscience; clone A7R34) and the geometric mean fluorescence intensity for IL-18R1, CD117, and CD127 staining on gated population was compared with respective isotype control staining intensities.
Flow cytometry
Freshly isolated thymocytes were stained as described above to discriminate the DN, DP, SP4, SP8, DN1-4, and the DN3/DN4 subsets (29) to establish gating parameters for cells harvested from OP9-DL4 co-cultures. A MoFlo-XDP cell sorter with Summit v4.3 software (Beckman Coulter, Fullerton, CA) was used for the experiments that included sorted populations. Cells were analyzed using a BD LSRII 4-laser flow cytometer and FACS Diva (BD Biosciences, San Jose, CA) and FlowJo software (Tree Star, Ashland, OR).
OP9-DL4 co-cultures
The OP9-DL4 cell line was kindly provided by Juan Carlos Zúñiga-Pflücker and maintained according to the protocols from his laboratory (30). For each experiment a fresh vial was thawed and grown to 60–80% confluence on treated plates; cells were then split and grown again to 60–80% confluence before the final plating on experimental 24-well treated plates. Sorted bone marrow and thymocyte subsets were co-cultured in plates with the OP9-DL4 stromal cells in αMEM (Invitrogen, NY) supplemented with 16.5% FBS (Sigma-Aldrich) and penicillin-streptomycin (Sigma-Aldrich) (culture media) and the cytokines indicated in the figures. It should be noted that no Flt3L was added to any of the cultures. After 7 days, the cells were harvested from the wells, counted, and stained for flow cytometry. Viable cell counts were obtained using 0.4% Trypan Blue (Lonza Inc, Allendale, NY) staining or annexin V and 7AAD staining technique.
Quantitative Real Time RT-PCR
Total RNA from sort-purified thymocyte subsets (2×104 cells) and splenic NK cells (1×105) were isolated using Qiagen (Germantown, MD) Mini-Elute columns. Total RNA was reverse transcribed to cDNA using a Qiagen Sensiscript Reverse Transcriptase kit. IL-18 receptor transcript abundance was measured by amplifying cDNA using IL-18Rα and IL-18Rβ primers from Quantitect (Qiagen) and quantified by the SYBR Green detection method using a ViiA™ 7 Real-Time PCR System (Life Technologies).
GAMMA (global microarray meta-analysis)
The GAMMA analysis predicted IL-18 should play a role in thymopoiesis and thymocyte differentiation based upon the genes it is highly correlated with across over 16,600 human microarray experiments. After identifying gene sets most specifically and consistently co-expressed with IL-18 across heterogeneous conditions, it used large-scale literature mining to identify what these co-expressed genes have in common. These commonalities become the inferred functions, roles and phenotypes for IL-18. Known functions serve as positive controls and, for IL-18, many of its major known roles were correctly predicted on the basis of its co-expressed genes, such as its pro-inflammatory role in the immune response and its ability to influence cytokine production, as well as genetic associations with other cytokines such as IL-12, IL-10, and IL-1.
Data Analysis
Flow cytometry data was analyzed using FACS Diva (BD Biosciences, San Jose, CA) and FlowJo software (Tree Star, Ashland, OR). Statistical analysis was performed using Graphpad Prism 6 Software and statistical significance between variables was estimated by performing One-way ANOVA and Fischer’s test for multiple comparisons.
Results
IL-18 acts in synergy with IL-7 to induce expansion of ETPs on OP9-DL4 stromal cells
An IL-18 dose response was performed in order to determine the effect of IL-18 on immature thymocytes in culture. Sort purified ETPs were cultured for one week on OP9-DL4 stroma with IL-7 added at a concentration of 5ng/ml in conjunction with IL-18 at concentrations ranging from 0.1 ng/ml to 100 ng/ml. Supplementing co-cultures with IL-18 significantly enhanced the expansion of ETP, as determined by total cell yields on day 7, compared to control treatments (Fig 1B). We observed that the magnitude of ETP expansion in the presence of IL-18 alone was comparable to the ETP expansion observed in the presence of IL-7 alone. Adding IL-7 and IL-18 together to the co-cultures greatly increased the cell yields when compared to cultures containing either IL-7 or IL-18 alone. The synergistic effects of IL-18 and IL-7 were evident only at higher doses of IL-18 (≥10ng/ml). Requirement of a higher IL-18 dose for cell response is not unusual as relatively higher concentrations of IL-18 are known to be required to activate cells in vitro (31). We next tested whether IL-18 influenced expansion of other immature thymocyte subsets including DN1d/e, DN2, and DN3 populations. We found that neither IL-7 nor IL-18 alone had an apparent effect on the expansion of these thymocytes, although co-cultures supplemented with both IL-7 and IL-18 showed a modest effect in promoting expansion of the DN2 population (Fig 1C). These results demonstrate that IL-18 can promote expansion of ETPs and can synergize with IL-7 in a dose dependent manner.
IL-7 and IL-18 stimulation enhance survival and proliferation of ETPs in OP9-DL4 co-cultures
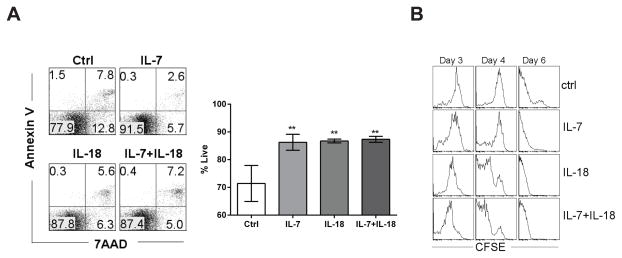
To assess whether increased cell yields in IL-7 and IL-18 stimulated ETP co-cultures were due to enhanced survival or increased proliferation, we measured cell viability in co-cultures by Annexin-V/7AAD staining and monitored cell divisions using a CFSE dilution assay. We observed that the percentage of live cells in ETP co-cultures stimulated with cytokines was significantly higher than that observed in unstimulated co-cultures (Figure 2A). IL-7 and IL-18 were equally potent in increasing live cell percentages in 7-day co-cultures. There was also no apparent synergistic effect in the IL-7 + IL-18 condition on cell survival, presumably because there was minimal cell death observed in these co-cultures stimulated with IL-7 or IL-18 alone. Because the differences in cell survival among the treatments are small, it’s unlikely that enhanced ETP expansion in stimulated cultures was entirely due to enhanced survival. Hence, we compared the proliferation kinetics of unstimulated and stimulated ETP co-cultures. CFSE profiles demonstrated that the ETPs had undergone more cell divisions than CFSE staining can reliable detect by day six, irrespective of the culture conditions (Figure 2B). However, CFSE profiles on day four clearly showed that ETPs stimulated with either IL-7 or IL-18 alone or in combination experienced more divisions compared to unstimulated ETP cultures. Importantly, the synergistic action of IL-7/IL-18 co-administration was observed on day 4. Together, these results support the contention that IL-18 promotes expansion of ETPs by enhancing both survival and proliferation of ETPs.
Figure 2. Enhanced proliferation and survival of ETP in the presence of IL-7 and IL-18.
Sort-purified ETPs were co-cultured with OP9-DL4 stromal cells in media supplemented with IL-18 or IL-7 alone or in combination. A) Survival of cells after 7 days of culture was assessed using Annexin V and 7AAD binding (percentages of cells in each quadrant are represented as the mean ± SEM of 3 replicates wells). B) CSFE dilution profiles for ETPs expanding under indicated conditions on day 3, 4, or 6. Histograms are representative of duplicate samples from each experimental condition and are representative of at least three experiments with similar results. **p<0.05
IL-18 accelerates the differentiation of immature thymocytes
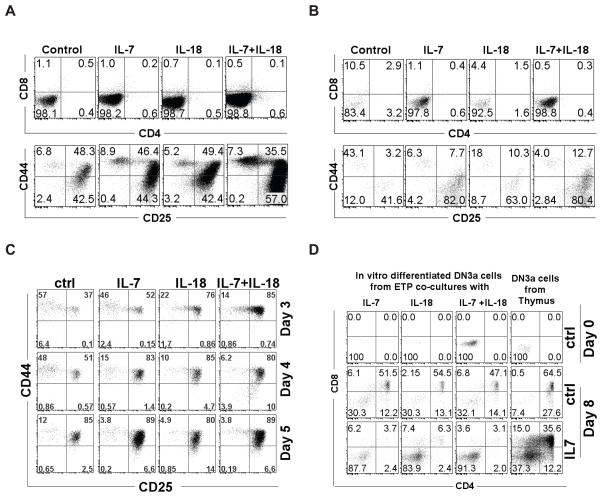
To determine whether IL-18 could influence the differentiation of immature thymocytes, sort-purified ETPs, or DN2s were co-cultured with OP9-DL4 stromal cells in the presence of IL-7 or IL-18 alone or in combination. After 7 days, differentiation of thymocytes in the co-cultures was analyzed by discriminating thymocyte populations using surface markers. Both ETP and DN2 co-cultures supplemented with recombinant IL-7 showed an increase in total cell number after 7 days without any notable changes in the percentages of different thymocyte populations in comparison to untreated co-cultures (Figures 3A and 3B). Similarly, ETP and DN2 co-cultures treated with either IL-18 alone or in combination with IL-7 showed an increase in total cell number without skewing the percentages of any particular thymocyte population as compared with IL-7 alone or untreated cultures. However when we evaluated differentiation of ETPs from CFSE dilution assays at earlier time points (Fig 3C), we saw that the differentiation of ETPs into DN2 and DN3 subsets occurred at a faster rate in IL-7 and IL-18-stimulated cultures compared to unstimulated controls. The CFSE profiles also showed no preferential expansion or differentiation of a particular thymocyte population in co-cultures stimulated in the presence of IL-18 (data not shown). To determine the capacity for further development along the T cell lineage, we evaluated DN3a thymocyte subsets generated in vitro from ETP/OP9-DL4 co-cultures and fresh ex vivo DN3a thymocytes sort-purified from the thymus for their potential to develop into DP cells. We found that all in vitro generated DN3a subsets, irrespective of the source and treatment conditions, differentiated into DP subsets when co-cultured on OP9-DL4 stromal cells for 7 days (Fig. 3D) in the absence of IL-7. However, in the presence of IL-7 there were significantly fewer DP thymocytes, as evidenced by the percentage of cells within the DP quadrants, from DN3a populations generated from ETP co-cultures as well as from DN3a populations from thymus. This effect is not surprising, as IL-7 has been shown to inhibit the transition of DN2/DN3 thymocytes to the DP stage (32). These results collectively suggest that IL-18 increased the expansion of immature thymocytes without interfering with their differentiation into more mature thymocytes.
Figure 3. IL-7 and IL-18 accelerate ETP differentiation without skewing to a particular subset.
Sort purified ETP or DN2 thymocytes were co-cultured with OP9-DL4 stromal cells in media supplemented with IL-18 or IL-7 alone or in combination. Differentiation of ETP and DN2 cells into more mature thymocytes were assessed by discriminating cells in the cultures using standard phenotypic markers. Thymocyte subsets identified in ETP/OP9-DL4 (A) or DN2/OP9-DL4 (B) co-cultures on day 7 under indicated conditions are shown. C) Thymocyte subsets identified in ETP/OP9-DL4 co-cultures during five-day expansion of ETPs under indicated conditions. D) DN3a thymocytes sorted from day 7 ETP or DN2 co-cultures or thymus from C57BL/6 mouse were further cultured for 8 days on OP9-DL4 stromal cells and their differentiation into SP4, SP8 or DP cells were analyzed using phenotypic markers. Dot plots were representative of 2–3 replicates in each treatment and each experiment was repeated at least three times with similar results.
ETP and DN2 subsets express IL-18 receptor transcript but not discernible levels of IL-18 receptor protein as assessed by flow cytometry
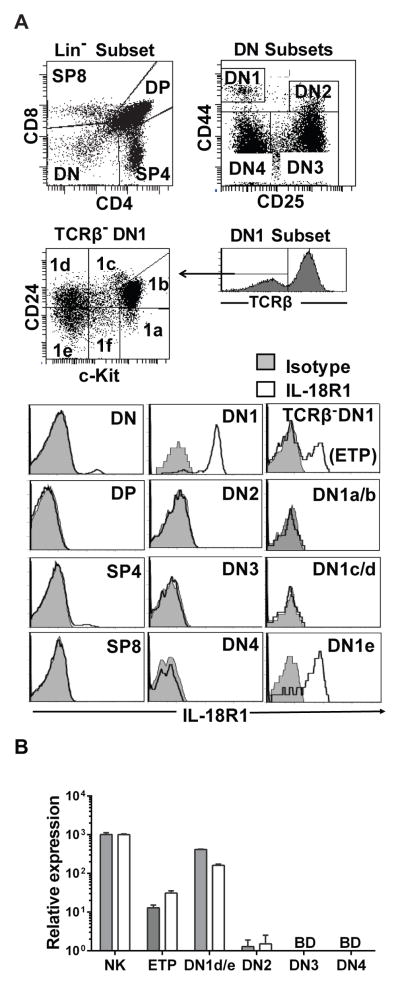
To further characterize the mechanism(s) by which IL-18 exerts its effect on developing T cells, surface expression of the IL-18R1 (CD218a) was assessed on the four main thymocyte populations, DN, DP, SP4 and SP8 (Figure 4A). Only a small percentage of the DN population and the SP4 population stained positively for the IL-18R1. Subdivision of the DN compartment revealed that only the DN1 population contained IL-18R1 expressing cells. Further parsing of the DN1 compartment by CD24 and c-Kit staining showed IL-18R1 surface expression to be restricted to the DN1e population. This is somewhat surprising given that the DN1a/b population (ETPs) expanded in the presence of IL-18 as did both ETPs and DN2 cells with the combination of both IL-7 and IL-18 as shown in Figure 1C.
Figure 4. The IL-18 receptor is differentially expressed on thymocyte subsets.
A) IL-18R1 protein expression evaluated by flow cytometry on freshly isolated thymocytes from C57BL/6 mouse. B) Real-time RT-PCR assessment of IL-18R1 (grey bar) and IL-18RAP (white bar) transcript abundance relative to GAPDH in sort-purified thymocyte subsets and NK cells from spleen. Data are representative of experiments repeated at least twice with similar results.
Because surface expression of the IL-18R1 was not detectible in the IL-18 responsive ETP and DN2 populations using flow cytometry, we assessed IL-18 receptor expression on ETP, DN1e/d, DN2, and DN3 thymocytes using Real-time RT-PCR. Both ETPs and DN2 expressed low levels of IL-18R1 and IL18Rap mRNA compared to DN1e/d populations and positive control splenic NK cells (Fig 4B). The OP9-DL4 stromal cells did not express IL-18 receptor either at transcript level or at surface expression (data not shown).
The IL-18 effect on thymocyte expansion is absent in cells from IL-18R1 deficient mice
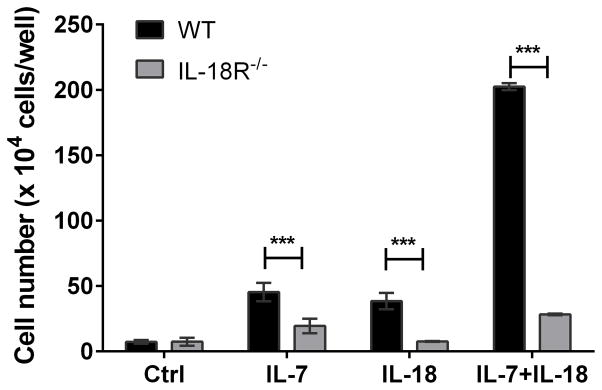
Although we did not detect surface expression of IL-18 receptors on ETPs or DN2 thymocytes, the presence of IL-18 receptor transcripts in these cells suggest the possibility for very low levels of receptor expression. To determine whether the IL-18 effects in the cultures were the direct result of IL-18 receptor engagement, we repeated the OP9-DL4 co-cultures described above comparing thymocytes from IL-18R1 null mice to wildtype thymocytes. ETPs from either C57/BL6 mice or IL-18R1 null mice (Figure 5) were plated with no cytokine, IL-7, IL-18, or a combination of IL-7 and IL-18. As expected, the IL-18R1 null mice did not show expansion in response to IL-18. The synergy between IL-18 and IL-7 was also absent in the null mice. This indicated that the IL-18 effects were mediated through the IL-18 receptor even though we were unable to detect IL-18R1 on the surface of the ETPs via flow cytometry.
Figure 5. The IL-18-induced increase in ETP expansion requires IL-18 receptor expression.
Sort purified ETPs from wildtype or IL-18R1−/− mouse thymocytes were co-cultured with OP9-DL4 stromal cells in media supplemented with IL-18 or IL-7 alone or in combination and cell yields from day 7 co-cultures were measured using a hemocytometer. Data are presented as means ± SEM of three replicate wells from each experimental condition and are representative of three experiments with similar results. ***p<0.001.
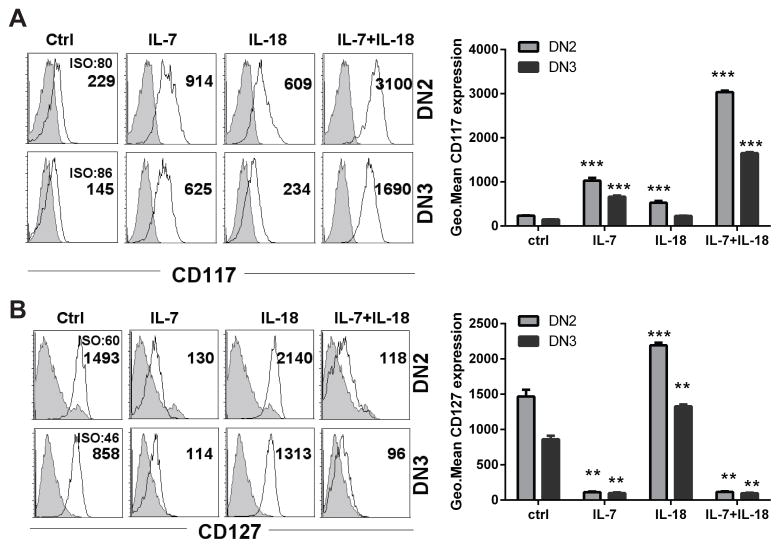
IL-18 stimulated ETPs significantly upregulate c-Kit and IL-7Rα receptor expression
C-Kit signaling has been shown to play an important role in promoting proliferation and differentiation of DN1 and DN2 thymocyte populations (33). Additionally, Zhoua et. al. (34) showed that recombinant IL-18 can positively regulate the expression of c-Kit in human melanocytes. To evaluate a potential mechanistic concurrence in our model, we tested the effects of IL-18 on regulation of c-Kit and IL-7Rα expression on immature thymocytes differentiating in the OP9-DL4 co-cultures. ETPs cultured in the presence of IL-7 or IL-18 alone for seven days demonstrated significantly upregulated c-Kit expression on the resulting DN2 and DN3 cells, though the effect was modest in comparison to the effect of adding both cytokines simultaneously (Figure 6). ETPs exposed to IL-7 plus IL-18 showed robust increases in surface c-Kit expression (~13.5-fold over control), which was significantly greater than what was observed in ETP cultures exposed to IL-7 or IL-18 alone (Figure 6A). Because IL-18 had a synergistic effect with IL-7, we also assessed the IL-7 receptor expression on these cells when treated with IL-18. As expected, stimulating ETPs with IL-18 caused a significant upregulation of surface IL-7Rα in differentiating thymocytes after seven days of co-culture (Figure 6B). ETP cultures exposed to IL-7 with or without IL-18 showed minimal IL-7Rα expression on the differentiating thymocytes, presumably due to activation-induced receptor internalization.
Figure 6. IL-18 induced upregulation of c-kit and IL-7Rα surface expression on ETP-OP9-DL4 co-cultures.
Sort purified ETPs from C57BL/6 mouse thymocytes were co-cultured for 7 days with OP9-DL4 stromal cells supplemented with IL-18 or IL-7 alone or in combination. DN2 and DN3 cell surface expression of (A) c-Kit (CD117) and (B) IL-7Rα (CD127) was analyzed by flow cytometry. Histograms are representative of three replicates in each treatment. Numbers in the histograms represent geometrical mean fluorescence intensity of CD117 or CD127 or their respective isotype control (ISO) staining. Bars represent mean ± SEM of receptor expression, as determined by fluorescence intensity, from 3 replicate wells in each treatment. Data are representative of three experiments with similar results. ***p<0.001.
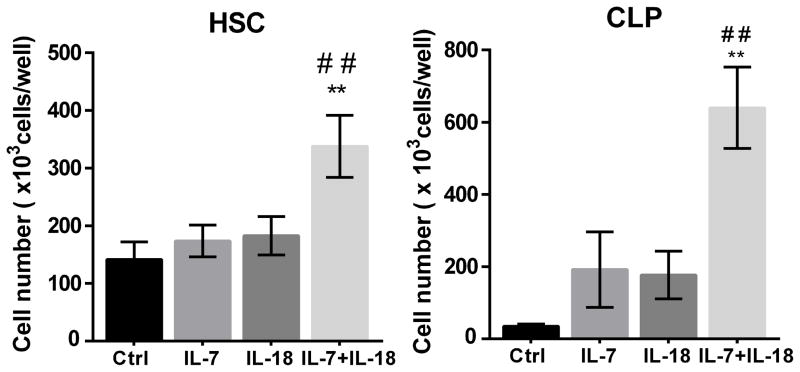
IL-18 promotes the expansion of immature progenitor cells in OP9-DL4 co-cultures
Lack of IL-18 mediated proliferative effects in DN1e/d despite IL-18 receptor expression by these cells suggested that IL-18 receptor expression alone is not sufficient for the proliferative effects. On the other hand, we found that cells responding to IL-18 both express c-kit and IL-7R, as well as upregulate these receptors after stimulation with IL-18. Hence, we hypothesized that the IL-18 proliferative effects are not restricted to immature thymocytes and may be present in other progenitor cells expressing c-kit and IL-7 receptors. To test this hypothesis we further investigated the proliferative effects of IL-18 on hematopoietic progenitor cells (HSC) and common lymphoid progenitor cells (CLP) isolated from mouse bone marrow, which also express the c-kit receptor. We observed that supplementing cultures with IL-7 and IL-18 significantly enhanced the proliferation of both HSC and CLP co-cultured on the OP9-DL4 stromal cells (Figure 7), although the proliferative effects of IL-7 and IL-18 combination is modest in co-cultures started with HSC compared to CLP.
Figure 7. IL-18 promotes expansion of HSCs and CLPs in OP9-DL4 co-cultures.
Sort purified HSC (1 ×104 cells) and CLP (500 cells) from C57BL/6 mouse bone marrow were co-cultured with OP9-DL4 stromal cells in media supplemented with IL-18 or IL-7 alone or in combination and cell yields from day 7 co-cultures were measured using a hemocytometer. Data are presented as means ± SEM of three replicate wells from each experimental condition and are representative of three experiments with similar results. ** Significant with p<0.05 compared to controls and ## significant with p<0.05 compared to IL-7 or IL-18.
Discussion
Humans have approximately 25,000 genes and, although their positions have been known since the completion of the draft genome in 2000, about one third of them still have yet to be characterized. Algorithmic approaches to predicting function on the basis of transcriptional network analysis, such as the GAMMA approach used here, enable function and phenotype to be predicted for genes (Figure 8). In the case of IL-18, much was already known; however, what we have shown here is that there can still be unknown/unexplored functions for known genes that can be inferred on the basis of their close neighbors in the transcriptional network.
Figure 8. The GAMMA principle.
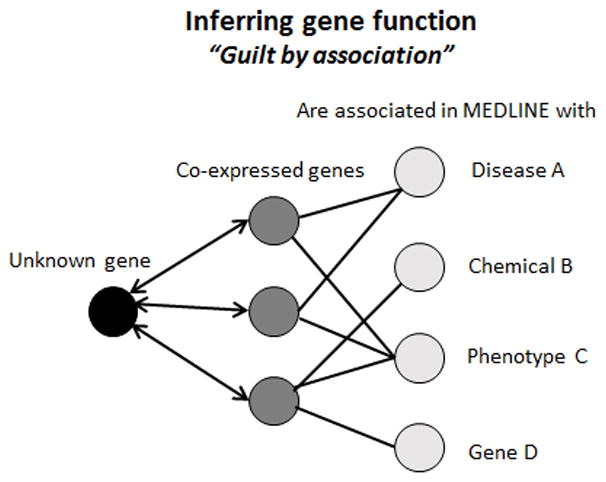
By using a set of 20 co-expressed (CoX) genes with known functions as a proxy, we can infer what an unknown gene does. Benchmark analyses using 5000 genes of known function confirm the approach is accurate. Experimental validations so far have corroborated the ability to predict function using this “guilt by association” approach.
As detailed in the Results section, IL-18 dramatically influenced T cell development in our in vitro model. IL-18 augmented thymocyte proliferation and accelerated differentiation capacity. Interestingly, the proliferative effects of IL-18 are not restricted to T cell progenitors in the thymus but also seen in more immature progenitor cells such as HSC, and CLPs in bone marrow that gives rise to all lymphoid-derived cells, suggesting the possibility that IL-18 can influence lymphopoiesis more broadly. Although IL-18 has well-established roles in a number of other processes, its involvement in early T cell development has not been well defined. IL-18 has been reported to be expressed within the microenvironment of the thymus (22) and therefore it is compelling to believe that it plays a functional role in T lymphopoiesis. IL-18 signaling-deficient mice demonstrate defects in peripheral Th1 responses, NK cell responses, and γδ T cell homing to lymph nodes (26, 35), although no gross defects in T cell development were reported in either IL-18-deficient or IL-18R1-deficient mice. The lack of a noticeable effect on T cell production (26, 35) suggests that the low level of IL-18 expressed in the thymus is not essential for T cell development under non-stressed conditions. However, the effects of this cytokine on thymocyte development could be missed unless the host system is stressed, as elevation of this cytokine would only be predicted to occur as a result of infection. Injection of IL-18 cDNA in conjunction with cDNAs for IL-2 or IL-12 in mice resulted in increased expression of cytokines (especially IFN-γ) from early thymocytes, giving at least indirect evidence that elevated levels of IL-18 can alter the thymic environment (24).
The IL-18-induced proliferative effects on thymocytes that we show here builds on previous evidence for a role for this cytokine in positive regulation of proliferation of T lymphocytes. This role was demonstrated in an earlier study (36) that showed IL-18 could induce proliferation of antigen-stimulated memory T cells in the periphery. In vivo relevance of the IL-18 effect was more recently reported in a study that showed IL-18 drove homeostatic expansion of naïve CD8 T cells in a lymphopenic host (37). The results we report here are intriguing because existing literature suggests that proinflammatory cytokines and inflammation in general negatively impact T lymphopoiesis (22–24,38, 39), which is in contrast with the IL-18 effects we have found on immature thymocytes and bone marrow progenitors. For instance, thymic involution has been shown to occur rapidly within 72 hr of endotoxin challenge (39). Although there is ample evidence suggesting that physiological stressors such as infection and inflammation can cause thymic atrophy due to apoptosis of DN and DP subsets (39, 40), the key players in restoring thymic homeostasis after infection or inflammation remain poorly defined. Because IL-18 showed effects similar to IL-7 in promoting proliferation and survival of ETPs, one could predict that IL-18 can act as a compensatory mechanism to boost thymic progenitors during an infection or inflammation and to restore thymic homeostasis. However intriguing, the complexities associated with exploring this hypothesis are beyond the scope of our current study.
Previous studies have demonstrated dendritic cell potential of DN1d and DN1e subsets in vivo (41) and the ability of IL-18 to drive differentiation of fetal DN1 cells to dendritic cells (22). However, in our OP9-DL4 co-cultures, IL-18- induced proliferative effects appeared restricted to DN1a/b (ETP) and DN2 cells that progressed toward DN3 as we observed no expansion of the of DN1d/e cells in this system (Figure 4). In any case, the experiments presented here clearly demonstrate that under controlled conditions, IL-18 can potentiate ETP proliferation and differentiation toward the T-lineage.
Notch signaling plays a crucial role in directing T cell development by tightly regulating various developmental steps including commitment, selection, proliferation, and survival of differentiating thymocytes (42). For instance, Notch can promote the proliferation and survival of DN populations by positively regulating the growth promoting signal pathways mediated by IL-7R and c-Kit (33, 43). There is ample evidence suggesting that both IL-7R and c-Kit play important roles in promoting the proliferation and survival of immature thymocytes (33, 44, 45). Consistent with these studies, we observed higher levels of both IL-7R and c-Kit on the surface of thymocytes expanded in the presence of IL-18 and IL-7 (Figure 6). These two receptor signaling pathways reportedly directly interact with each other (46) and positively regulate STAT5 signaling. Hence, it is plausible that the IL-18 effects in augmenting thymocyte expansion are mediated through positive regulation of a c-Kit and IL-7 signaling axis. However, further studies are essential to confirm this contention and to explore whether IL-18 modulates c-kit and IL-7R expression directly using transcriptional or post-transcriptional mechanisms or indirectly by modulating notch signaling, which is known to modulate the expression of these receptors on the surface.
Perhaps most importantly, we have demonstrated that IL-18 can substitute for IL-7 in early thymic development processes and can synergize with IL-7 in promoting proliferation. This finding warrants further investigation to determine whether co-administration of IL-18 and IL-7 will be valuable for therapeutic purposes. Administration of recombinant IL-7 has shown promise in clinical trials due to its ability to promote lymphopoiesis under lymphopenic conditions (47, 48). However, use of IL-7 has limitations such as its bias towards homeostatic expansion of peripheral lymphocytes and its marginal effects on enhancing progenitor cell populations that give rise to lymphocyte diversity(49, 50). Furthermore, the duration and dose of IL-7 necessary for effect pose a risk of graft versus host disease (GVHD) due to its ability to enhance T cell functions (48, 51). IL-18 has also been put into the spotlight for its potential role in cancer treatment and is showing promising results in pre-clinical and clinical studies (52–54). Based on the robust synergistic effects observed in our assays and the recently reported effects of IL-18 in driving homeostatic expansion of naïve CD8 T cells in lymphopenic mice (37), it is possible that combining IL-18 and IL-7 as a combination therapy in humans could overcome the limitations of IL-7, by enhancing the IL-7 effects on bone marrow and thymus progenitor cells and decreasing the dose and duration of IL-7 needed for lymphocyte reconstitution in clinical scenarios of lymphodepletion. Alternatively, this synergy could be exploited to expand progenitor cells ex vivo for reconstitution into lymphopenic hosts.
As IL-7 has been characterized to be absolutely required for “normal” T cell development, it will be interesting to further characterize and compare the differences in phenotypically identical cells that have been raised in IL-7 versus IL-18 environments. The value of these studies lies in the ability to potentially co-opt these signaling pathways for future potential therapeutic purposes by enhancing our ability to elaborate T cells in vivo following lymphodepletion or in vitro for envisioned adoptive immunotherapy or reconstitution efforts.
Acknowledgments
The BD LSR II flow cytometer was a gift from the Oxley Foundation. We thank the Chapman Foundation for providing seed funding to begin this work, as well as the NIH (grants # 5P20GM103636 and 8P20GM103456 to J.D.W.), the Oklahoma Center for the Advancement of Science and Technology (grant #HR07-095 to T.K.T.), and the University of Oklahoma School of Community Medicine, Integrative Immunology Center for additional funding.
Grant Support: Support for this project was received from the Chapman Foundation, the Oklahoma Medical Research Foundation, the Oklahoma Center for the Advancement of Science and Technology #HR07-095(TKT), the NIH#5P20RR020143-07 (JDW) and the University of Oklahoma School of Community Medicine, Integrative Immunology Center.
Abbreviations
- DL
Delta-like
- DN
Double Negative
- DP
Double Positive Thymocyte
- ETPs
Early Thymic Progenitors
- GAMMA
Global Microarray Meta-Analysis
- IL18RAP
IL-18 Receptor Accessory Protein
- IRIDESCENT
Implicit Relationship IDEntification by in-Silico Construction of an Entity based Network from Text
- ISP
Immature Single Positive
- SP
Single Positive Thymocyte
References
- 1.Wren JD. A global meta-analysis of microarray expression data to predict unknown gene functions and estimate the literature-data divide. Bioinformatics. 2009;25:1694–1701. doi: 10.1093/bioinformatics/btp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wren JD. Extending the mutual information measure to rank inferred literature relationships. BMC bioinformatics. 2004;5:145. doi: 10.1186/1471-2105-5-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wren JD, Bekeredjian R, Stewart JA, Shohet RV, Garner HR. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics. 2004;20:389–398. doi: 10.1093/bioinformatics/btg421. [DOI] [PubMed] [Google Scholar]
- 4.Wren JD, Garner HR. Shared relationship analysis: ranking set cohesion and commonalities within a literature-derived relationship network. Bioinformatics. 2004;20:191–198. doi: 10.1093/bioinformatics/btg390. [DOI] [PubMed] [Google Scholar]
- 5.Daum JR, Wren JD, Daniel JJ, Sivakumar S, McAvoy JN, Potapova TA, Gorbsky GJ. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Current biology: CB. 2009;19:1467–1472. doi: 10.1016/j.cub.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Towner RA, Jensen RL, Colman H, Vaillant B, Smith N, Casteel R, Saunders D, Gillespie DL, Silasi-Mansat R, Lupu F, Giles CB, Wren JD. ELTD1, a potential new biomarker for gliomas. Neurosurgery. 2013;72:77–90. doi: 10.1227/NEU.0b013e318276b29d. discussion 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemmensen SN, Bohr CT, Rorvig S, Glenthoj A, Mora-Jensen H, Cramer EP, Jacobsen LC, Larsen MT, Cowland JB, Tanassi JT, Heegaard NH, Wren JD, Silahtaroglu AN, Borregaard N. Olfactomedin 4 defines a subset of human neutrophils. Journal of leukocyte biology. 2012;91:495–500. doi: 10.1189/jlb.0811417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lupu C, Zhu H, Popescu NI, Wren JD, Lupu F. Novel protein ADTRP regulates TFPI expression and function in human endothelial cells in normal conditions and in response to androgen. Blood. 2011;118:4463–4471. doi: 10.1182/blood-2011-05-355370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Towner RA, Jensen RL, Vaillant B, Colman H, Saunders D, Giles CB, Wren JD. Experimental validation of 5 in-silico predicted glioma biomarkers. Neuro-oncology. 2013;15:1625–1634. doi: 10.1093/neuonc/not124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamura H, Tsutsi H, Komatsu T, Yutsudo M, Hakura A, Tanimoto T, Torigoe K, Okura T, Nukada Y, Hattori K, et al. Cloning of a new cytokine that induces IFN-gamma production by T cells. Nature. 1995;378:88–91. doi: 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- 11.Gracie JA, Robertson SE, McInnes IB. Interleukin-18. Journal of leukocyte biology. 2003;73:213–224. doi: 10.1189/jlb.0602313. [DOI] [PubMed] [Google Scholar]
- 12.Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunological reviews. 2008;223:20–38. doi: 10.1111/j.1600-065X.2008.00624.x. [DOI] [PubMed] [Google Scholar]
- 13.Dao T, Ohashi K, Kayano T, Kurimoto M, Okamura H. Interferon-gamma-inducing factor, a novel cytokine, enhances Fas ligand-mediated cytotoxicity of murine T helper 1 cells. Cellular immunology. 1996;173:230–235. doi: 10.1006/cimm.1996.0272. [DOI] [PubMed] [Google Scholar]
- 14.Okamoto I, Kohno K, Tanimoto T, Ikegami H, Kurimoto M. Development of CD8+ effector T cells is differentially regulated by IL-18 and IL-12. Journal of immunology. 1999;162:3202–3211. [PubMed] [Google Scholar]
- 15.Komai-Koma M, Gracie JA, Wei XQ, Xu D, Thomson N, McInnes IB, Liew FY. Chemoattraction of human T cells by IL-18. Journal of immunology. 2003;170:1084–1090. doi: 10.4049/jimmunol.170.2.1084. [DOI] [PubMed] [Google Scholar]
- 16.Hoshino T, Winkler-Pickett RT, Mason AT, Ortaldo JR, Young HA. IL-13 production by NK cells: IL-13-producing NK and T cells are present in vivo in the absence of IFN-gamma. Journal of immunology. 1999;162:51–59. [PubMed] [Google Scholar]
- 17.Nakanishi K, Yoshimoto T, Tsutsui H, Okamura H. Interleukin-18 is a unique cytokine that stimulates both Th1 and Th2 responses depending on its cytokine milieu. Cytokine & growth factor reviews. 2001;12:53–72. doi: 10.1016/s1359-6101(00)00015-0. [DOI] [PubMed] [Google Scholar]
- 18.Yoshimoto T, Mizutani H, Tsutsui H, Noben-Trauth N, Yamanaka K, Tanaka M, Izumi S, Okamura H, Paul WE, Nakanishi K. IL-18 induction of IgE: dependence on CD4+ T cells, IL-4 and STAT6. Nature immunology. 2000;1:132–137. doi: 10.1038/77811. [DOI] [PubMed] [Google Scholar]
- 19.Gutcher I, Urich E, Wolter K, Prinz M, Becher B. Interleukin 18-independent engagement of interleukin 18 receptor-alpha is required for autoimmune inflammation. Nature immunology. 2006;7:946–953. doi: 10.1038/ni1377. [DOI] [PubMed] [Google Scholar]
- 20.Finotto S, Siebler J, Hausding M, Schipp M, Wirtz S, Klein S, Protschka M, Doganci A, Lehr HA, Trautwein C, Khosravi-Far R, Strand D, Lohse A, Galle PR, Blessing M, Neurath MF. Severe hepatic injury in interleukin 18 (IL-18) transgenic mice: a key role for IL-18 in regulating hepatocyte apoptosis in vivo. Gut. 2004;53:392–400. doi: 10.1136/gut.2003.018572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoshino T, Kawase Y, Okamoto M, Yokota K, Yoshino K, Yamamura K, Miyazaki J, Young HA, Oizumi K. Cutting edge: IL-18-transgenic mice: in vivo evidence of a broad role for IL-18 in modulating immune function. Journal of immunology. 2001;166:7014–7018. doi: 10.4049/jimmunol.166.12.7014. [DOI] [PubMed] [Google Scholar]
- 22.Ito H, Esashi E, Akiyama T, Inoue J, Miyajima A. IL-18 produced by thymic epithelial cells induces development of dendritic cells with CD11b in the fetal thymus. International immunology. 2006;18:1253–1263. doi: 10.1093/intimm/dxl058. [DOI] [PubMed] [Google Scholar]
- 23.Youm YH, Kanneganti TD, Vandanmagsar B, Zhu X, Ravussin A, Adijiang A, Owen JS, Thomas MJ, Francis J, Parks JS, Dixit VD. The Nlrp3 inflammasome promotes age-related thymic demise and immunosenescence. Cell reports. 2012;1:56–68. doi: 10.1016/j.celrep.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez-Galan MC, Bream JH, Farr A, Young HA. Synergistic effect of IL-2, IL-12, and IL-18 on thymocyte apoptosis and Th1/Th2 cytokine expression. Journal of immunology. 2005;174:2796–2804. doi: 10.4049/jimmunol.174.5.2796. [DOI] [PubMed] [Google Scholar]
- 25.Holmes R, Zuniga-Pflucker JC. The OP9-DL1 system: generation of T-lymphocytes from embryonic or hematopoietic stem cells in vitro. Cold Spring Harbor protocols. 2009;2009 doi: 10.1101/pdb.prot5156. pdb prot5156. [DOI] [PubMed] [Google Scholar]
- 26.Hoshino K, Tsutsui H, Kawai T, Takeda K, Nakanishi K, Takeda Y, Akira S. Cutting edge: generation of IL-18 receptor-deficient mice: evidence for IL-1 receptor-related protein as an essential IL-18 binding receptor. Journal of immunology. 1999;162:5041–5044. [PubMed] [Google Scholar]
- 27.Teague TK, Schaefer BC, Hildeman D, Bender J, Mitchell T, Kappler JW, Marrack P. Activation-induced inhibition of interleukin 6-mediated T cell survival and signal transducer and activator of transcription 1 signaling. The Journal of experimental medicine. 2000;191:915–926. doi: 10.1084/jem.191.6.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van de Wiele CJ, Marino JH, Tan C, Kneale HA, Weber J, Morelli JN, Davis BK, Taylor AA, Teague TK. Impaired thymopoiesis in interleukin-7 receptor transgenic mice is not corrected by Bcl-2. Cellular immunology. 2007;250:31–39. doi: 10.1016/j.cellimm.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan C, Taylor AA, Coburn MZ, Marino JH, Van De Wiele CJ, Teague TK. Ten-color flow cytometry reveals distinct patterns of expression of CD124 and CD126 by developing thymocytes. BMC immunology. 2011;12:36. doi: 10.1186/1471-2172-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 31.Dinarello CA, Novick D, Kim S, Kaplanski G. Interleukin-18 and IL-18 Binding Protein. Frontiers in immunology. 2013;4:289. doi: 10.3389/fimmu.2013.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang J, Garrett KP, Pelayo R, Zuniga-Pflucker JC, Petrie HT, Kincade PW. Propensity of adult lymphoid progenitors to progress to DN2/3 stage thymocytes with Notch receptor ligation. Journal of immunology. 2005;175:4858–4865. doi: 10.4049/jimmunol.175.8.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massa S, Balciunaite G, Ceredig R, Rolink AG. Critical role for c-kit (CD117) in T cell lineage commitment and early thymocyte development in vitro. European journal of immunology. 2006;36:526–532. doi: 10.1002/eji.200535760. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J, Shang J, Song J, Ping F. Interleukin-18 augments growth ability of primary human melanocytes by PTEN inactivation through the AKT/NF-kappaB pathway. The international journal of biochemistry & cell biology. 2013;45:308–316. doi: 10.1016/j.biocel.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Takeda K, Tsutsui H, Yoshimoto T, Adachi O, Yoshida N, Kishimoto T, Okamura H, Nakanishi K, Akira S. Defective NK cell activity and Th1 response in IL-18-deficient mice. Immunity. 1998;8:383–390. doi: 10.1016/s1074-7613(00)80543-9. [DOI] [PubMed] [Google Scholar]
- 36.Iwai Y, Hemmi H, Mizenina O, Kuroda S, Suda K, Steinman RM. An IFN-gamma-IL-18 signaling loop accelerates memory CD8+ T cell proliferation. PloS one. 2008;3:e2404. doi: 10.1371/journal.pone.0002404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walsh MC, Pearce EL, Cejas PJ, Lee J, Wang LS, Choi Y. IL-18 Synergizes with IL-7 To Drive Slow Proliferation of Naive CD8 T Cells by Costimulating Self-Peptide-Mediated TCR Signals. Journal of immunology. 2014 doi: 10.4049/jimmunol.1400396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Billard MJ, Gruver AL, Sempowski GD. Acute endotoxin-induced thymic atrophy is characterized by intrathymic inflammatory and wound healing responses. PloS one. 2011;6:e17940. doi: 10.1371/journal.pone.0017940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang SD, Huang KJ, Lin YS, Lei HY. Sepsis-induced apoptosis of the thymocytes in mice. Journal of immunology. 1994;152:5014–5021. [PubMed] [Google Scholar]
- 40.Gruver AL, Sempowski GD. Cytokines, leptin, and stress-induced thymic atrophy. Journal of leukocyte biology. 2008;84:915–923. doi: 10.1189/jlb.0108025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore AJ, Sarmiento J, Mohtashami M, Braunstein M, Zuniga-Pflucker JC, Anderson MK. Transcriptional priming of intrathymic precursors for dendritic cell development. Development. 2012;139:373–384. doi: 10.1242/dev.069344. [DOI] [PubMed] [Google Scholar]
- 42.Shah DK, Zuniga-Pflucker JC. Notch receptor-ligand interactions during T cell development, a ligand endocytosis-driven mechanism. Current topics in microbiology and immunology. 2012;360:19–46. doi: 10.1007/82_2012_225. [DOI] [PubMed] [Google Scholar]
- 43.Gonzalez-Garcia S, Garcia-Peydro M, Alcain J, Toribio ML. Notch1 and IL-7 receptor signalling in early T-cell development and leukaemia. Current topics in microbiology and immunology. 2012;360:47–73. doi: 10.1007/82_2012_231. [DOI] [PubMed] [Google Scholar]
- 44.von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SE, Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. The Journal of experimental medicine. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maki K, Sunaga S, Komagata Y, Kodaira Y, Mabuchi A, Karasuyama H, Yokomuro K, Miyazaki JI, Ikuta K. Interleukin 7 receptor-deficient mice lack gammadelta T cells. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:7172–7177. doi: 10.1073/pnas.93.14.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jahn T, Sindhu S, Gooch S, Seipel P, Lavori P, Leifheit E, Weinberg K. Direct interaction between Kit and the interleukin-7 receptor. Blood. 2007;110:1840–1847. doi: 10.1182/blood-2005-12-028019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capitini CM, Chisti AA, Mackall CL. Modulating T-cell homeostasis with IL-7: preclinical and clinical studies. Journal of internal medicine. 2009;266:141–153. doi: 10.1111/j.1365-2796.2009.02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sasson SC, Zaunders JJ, Kelleher AD. The IL-7/IL-7 receptor axis: understanding its central role in T-cell homeostasis and the challenges facing its utilization as a novel therapy. Current drug targets. 2006;7:1571–1582. doi: 10.2174/138945006779025365. [DOI] [PubMed] [Google Scholar]
- 49.Chu YW, Memon SA, Sharrow SO, Hakim FT, Eckhaus M, Lucas PJ, Gress RE. Exogenous IL-7 increases recent thymic emigrants in peripheral lymphoid tissue without enhanced thymic function. Blood. 2004;104:1110–1119. doi: 10.1182/blood-2003-10-3635. [DOI] [PubMed] [Google Scholar]
- 50.Broers AE, Posthumus-van Sluijs SJ, Spits H, van der Holt B, Lowenberg B, Braakman E, Cornelissen JJ. Interleukin-7 improves T-cell recovery after experimental T-cell-depleted bone marrow transplantation in T-cell-deficient mice by strong expansion of recent thymic emigrants. Blood. 2003;102:1534–1540. doi: 10.1182/blood-2002-11-3349. [DOI] [PubMed] [Google Scholar]
- 51.Goldberg GL, Zakrzewski JL, Perales MA, van den Brink MR. Clinical strategies to enhance T cell reconstitution. Seminars in immunology. 2007;19:289–296. doi: 10.1016/j.smim.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srivastava S, Salim N, Robertson MJ. Interleukin-18: biology and role in the immunotherapy of cancer. Current medicinal chemistry. 2010;17:3353–3357. doi: 10.2174/092986710793176348. [DOI] [PubMed] [Google Scholar]
- 53.Tarhini AA, Millward M, Mainwaring P, Kefford R, Logan T, Pavlick A, Kathman SJ, Laubscher KH, Dar MM, Kirkwood JM. A phase 2, randomized study of SB-485232, rhIL-18, in patients with previously untreated metastatic melanoma. Cancer. 2009;115:859–868. doi: 10.1002/cncr.24100. [DOI] [PubMed] [Google Scholar]
- 54.Simpkins F, Flores A, Chu C, Berek JS, Lucci J, 3rd, Murray S, Bauman J, Struemper H, Germaschewski F, Jonak Z, Gardner O, Toso J, Coukos G. Chemoimmunotherapy using pegylated liposomal Doxorubicin and interleukin-18 in recurrent ovarian cancer: a phase I dose-escalation study. Cancer immunology research. 2013;1:168–178. doi: 10.1158/2326-6066.CIR-13-0098. [DOI] [PubMed] [Google Scholar]