Abstract
Heat shock factor 1 (HSF1) has long been recognized as the master transcription factor that regulates heat shock proteins (HSPs). More recently HSF1 has been associated with a broader role in regulating response to a variety of cellular stresses beyond heat-shock. We previously found that high HSF1 expression is associated with poor outcome in lung, breast and colon cancers. Importantly, however, the HSF1 signature correlated with poor outcome in these studies was not related to the heat shock response, which suggested that tumor outcome associated with high HSF expression may be due to processes other than stress response. Hence, we explored the question whether high HSF1 expression might be associated with the cancer stem cell (CSC) phenotype. To do so, we examined the association of HSF1 with CSC phenotype by FACS and immunofluorescence. In addition, we evaluated the effects of HSF1 over-expression and knock-down on sphere formation and CSC marker expression in breast cancer cell lines. Here, we report results demonstrating that high HSF1 not only correlates with CSC marker expression, but inducible HSF1 over-expression augments and HSF1 knock-down inhibits CSC phenotype. Furthermore, HSF1 expression confers resistance to chemotherapeutic drugs and increases CSC frequency. In conclusion, our study indicates that one of the potential HSP-independent HSF1 driven mechanisms that may contribute to poor outcome in human tumors involves regulation of the CSC phenotype. Hence, therapeutic inhibition of HSF1 may be one route to target CSCs in human tumors.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-015-3521-1) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Heat shock factor 1 (HSF1), Cancer stem cell, Tumorsphere assay
Introduction
Cancer stem cells (CSCs) are subpopulation of tumor cells that are capable of self-renewal and differentiation [1–4]. CSCs have the ability to initiate tumors with extremely low seeding numbers in immunocompromised mice and differentiate asymmetrically into non-CSCs while maintaining a pool of CSCs. It is thought that CSCs may be resistant to chemotherapy and radiation due to their low rate of cell division [5].
While the presence of CSCs has been documented in multiple human tumors, the mechanisms that regulate CSCs remain to be fully elucidated. Several pathways including Hedgehog, NOTCH, and Wnt/β-catenin signaling have been associated with the CSC phenotype [6–8]. However, the rare nature of CSCs coupled with the methodological challenges associated with isolating, maintaining, and expanding them in vitro has slowed progress. Hence, much remains to be learned about CSC biology.
Heat shock factor 1 (HSF1) is the master transcription regulator of heat shock response (HSR). In addition, HSF1 coordinates the response of cells to diverse arrays of environmental and physiological stresses, such as ischemic injury, heavy metals, chemicals, free-radicals, inflammation, etc. [9].
We recently demonstrated that HSF1 is highly expressed in multiple human tumors including lung, colon, breast, and ovarian cancers. Importantly, these studies showed that HSF1 over-expression is associated with significantly worse outcome in these tumors [10, 11]. Through genome-wide ChIP-Seq analysis, we also discovered that HSF1 regulates two classes of genes in tumor cells; one set of genes included well-known stress response related targets such as HSPs; however, a second set of genes were involved with various other cellular processes, including cell cycle, signaling, metabolism, adhesion, and protein translation [11]. Somewhat surprisingly, it was the non-heat shock HSF1 target genes that were closely associated with the more aggressive tumor phenotype, poor outcome, metastasis, and death in breast, colon, and lung tumors isolated directly from patients [11]. High HSF1 expression has also been associated with poor outcome, advanced stage or metastasis in endometrial Carcinoma, hepatocellular carcinoma, melanoma, and squamous cell carcinoma [12–15].
In the present study, we provide evidence that the heat shock-independent activation of HSF1 augments the breast cancer stem cell phenotype. These results provide a new mechanism for the association between high HSF1 expression and poor outcome observed in human tumors.
Materials and methods
Cell culture
BPLER and HMLER cells were described previously [16] and obtained from live tumor culture core (LTCC, http://sylvester.org/shared-resources/Live-Tumor-Culture-Core). In brief, the BPLER cells are cultured in BMI-T medium obtained from LTCC, and HMLER cells are cultured in MEGM medium (Lonza, Cat. No. CC-3150). The numbers subsequent to the cell name (i.e., 2, 3, and 4) indicate cell lines established from different patient donors. SUM159 and SUM190 were kindly provided by Dr. Stephen Ethier and cultured in their recommended media (Ham’s F12 contains 5 % fetal bovine serum, 5 µg/ml insulin, and 1 µg/ml hydrocortisone). All other cell lines were obtained from the American Type Culture Collection (ATCC) and cultured in recommended conditions. All the cell lines were validated by STR analysis and tested negative for mycoplasma.
Real-time RT-PCR
Quantitative real-time RT-PCR was performed using SuperScript® III First-Strand Synthesis kit and SsoFast ™ EvaGreen® Supermix according to the manufacturers’ instructions and analyzed with BioRad CFX Manager Software. GAPDH mRNA was used as an internal control to normalize RNA inputs, and expression levels were calculated according to the relative ΔCt method. Primers sequences are annotated in Supplemental Table 1.
Flow cytometry and cell sorting
Cells were harvested at 80 % confluence with non-enzymatic dissociation buffer (Life Technologies, Cat. No. 13151-014) and cell pellets were washed, resuspended in Hank’s Balanced Salt Solution (HBSS) buffer containing 0.2 % BSA, and stained with fluorophore-conjugated monoclonal antibodies per manufacture’s instruction. Antibodies used for cell sorting included anti-CD44, APC conjugated (BD Pharmingen, 559942) and CD166, PE conjugated (BD Pharmingen, 559263). Flow sorting was performed using BD FACS Aria II and purity of sorted cells was confirmed to be between 90-95 %. Sorted cells were washed once with HBSS before protein extraction.
Immunoflurescence staining
Cells were plated on 8-chamber slides, fixed with 4 % PFA for 15 min at room temperature, and then permeabilized for 10 min with 0.2 % Tween-20. After blocking for 1 h in 10 % Goat serum, slides were incubated overnight with primary antibodies at 4 °C, followed by 45 min incubation with Alexa Fluorophore-labeled secondary antibodies. Fluoro-Gel II containing DAPI (Electron Microscopy Sciences, Cat. No. 17985-50) was used to mount slides, and images were taken on an EVOS fluorescence microscope (AMG & Life Tech). Primary antibodies and their dilutions used in our experiments are listed in Supplemental Table 2.
Doxycycline inducible HSF1 over-expression and knockdown
For inducible HSF1 over-expression, a complete HSF1 ORF was subcloned into pENTR1A gateway entry vector (ENTR) and transferred into pLenti CMV/TO Hygro DEST plasmid (Addgene, 17291) provided by Dr. Eric Campeau [17]. pLenti CMV TetR Blast was used for Tet-repressor expression (Addgene 17492). For inducible HSF1 knockdown, specific HSF1 shRNA sequences were obtained from Dr. Susan Lindquist [11] and cloned into pLKO-Tet-On-Neo plasmid provided by Dr. Dmitri Wiederschain from Addgene (Cat. No. 21916) [18]. Non-target shRNA (NT shRNA) was cloned into the same plasmid to serve as a control. Lentiviral packaging was achieved using the second generation packaging plasmid pCMV-dR8.2 dvpr and pCMV-VSV-G (Addgene Cat. No. 8455 and 8454). Lentivirus-infected cells were selected with antibiotic treatment. Doxycycline (DOX) was added into cell culture to induce HSF1 over-expression or knockdown before experiments. To ensure that the experiments were carried out below toxic DOX doses, first serial dilutions of DOX were tested and 1 ng/ml was determined to be optimal for use in HSF1 over-expression or knockdown.
Tumorsphere formation
Tumorsphere formation assays were performed following Clarke et al. with slight modification [19]. In brief, 3000 to 5000 cells were plated on 6-well plates pretreated with polyhema and maintained in 4-ml tumorsphere media. The tumorsphere medium is composed of a 1:1 ratio of DMEM/F12 containing 2 % B27 (Invitrogen), 20 ng/ml EGF, 20 ng/ml bFGF (BD Biosciences), 4 µg/ml heparin (Sigma), and 0.5 % methyl cellulose. For inducible HSF1 over-expression or knockdown groups, cells were pretreated with DOX for 48 h before sphere assay and sphere medium was supplemented with DOX to maintain the induction. After 10–14 days of incubation, spheres were consolidated into a 96-well plate and imaged on an EVOS fluorescence microscope (AMG & Life Tech) after DAPI staining and they were also counted using GelCount™ (Oxford Optronix) following iodonitratetrazolium chloride staining. Each group included triplicate samples and at least 3 independent experiments were carried out.
Western blotting
Western blot assays were performed as described before [20]. Protein lysates were harvested with RIPA buffer and run on SDS-PAGE gels. Blots were probed with antibodies listed in Table S2. Blots were developed using SuperSignal West regular or Dura ECL (ThermoScientific).
Drug treatments
The effect of paclitaxel (Taxol) on cell proliferation was tested both in 2D and 3D cultures. For 2D culture, cells were treated with Taxol at 2.5 nM with HSF1 over-expression or knockdown for 4 days and counted with Trypan blue exclusion method using a Nexcelom Auto T4 Cellometer. For 3D culture, 5 nM taxol was added into pre-induced HSF1 over-expression or knockdown cells and maintained for 4 days. Cells were allowed to recover for 3 days before trypsinization and subjected to tumorsphere assay. Percentage inhibition on 2D or 3D cell culture was compared to control groups which only had vehicle addition.
For protein translation inhibition, 3 pairs of HMLER and BPLER cells were plated at 8000 cell/well on 96-well plate one day before adding protein translation inhibitors including anisomycin and cycloheximide. Protein translation inhibitor drugs were removed after 8 h incubation and cells were maintained in fresh drug-free media for additional four days before being subjected to CellTiter-Blue® (Promega, Cat. No. G8081) assay. Inhibition of cell proliferation inhibition was compared to the vehicle control group.
Statistical analysis
Inhibition of cell proliferation and sphere formation was analyzed using student’s t test or one-way ANOVA with significance at p < 0.05.
Results
HSF1 is highly expressed in CSCs
We initially explored the potential role of HSF1 in CSCs using CSC-like BPLER and non-CSC-like HMLER cell lines. Both BPLER and HMLER lines do form mouse xenograft tumors that are histologically similar to human primary triple-negative breast carcinoma (TNBC), and their global mRNA profile mimics human TNBC [21]. We previously demonstrated that only the BPLER cells possessed CSC-like features, even though the matched HMLER cell lines are isolated from the same patient and transformed with identical oncogenes [16, 20]. In vivo tumor formation studies showed that as low as ten unsorted BPLER cells could form tumors in mice. In contrast, >104 HMLER cells were required for tumor formation. Consistent with these observations, we now show that BPLER cells express high levels of CSC-associated markers such as CD326 (EpCAM), CD44v, and CD166 and form fivefold more tumor spheres compared to HMLER (Fig. 1a) [22].
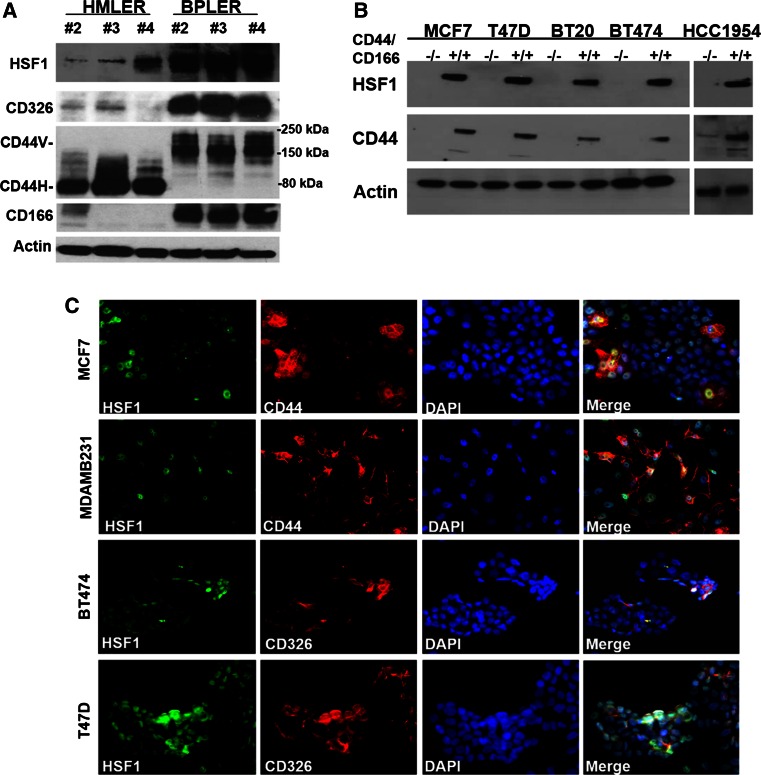
Fig. 1.
High HSF1 expression is associated with CSCs in multiple cell lines. a HSF1 is highly expressed in CSC-like BPLER cells that express CSC-associated markers CD326, high-molecular weight CD44v, and CD166 compared to isogenic non-CSC-like HMLER cells with low CSC marker expression. Western blot of three matched pairs (#2, 3, and 4) of HMLER and BPLER cell lines was performed with antibodies against HSF1, CD326, CD44, and CD166. β-actin (Actin) was used as loading control. CD44H: CD44 standard form. b HSF1 is highly expressed in FACS-enriched CD44high/CD166high CSC population (+/+) compared to CD44low/CD166low non-CSC population (−/−) in multiple breast cancer cell lines; MCF7, T47D, BT20, BT474, and HCC1954. Western blot of protein lysates were performed with anti-HSF1 and CD44 antibodies, β-actin (Actin) was used as loading control. c Double immunofluorescence demonstrates that HSF1 and CSC markers are co-expressed in the same subpopulation of cells in situ in multiple breast cancer cell lines; MCF7, MDAMB231, BT474, and T47D. The cells were plated in 8-chamber slides and simultaneously probed with HSF1 (green) and CSC antibodies CD44 or CD326 (red), and overlaid with nuclear DAPI stain (blue) and images were captured with an EVOS fluorescence microscope at 20X magnification
Taking the advantage of this cell model system, we compared HSF1 expression between BPLER and HMLER cells using real-time PCR and Western blotting, and found that HSF1 is over-expressed in all three CSC-like BPLER cell lines compared to non-CSC-like HMLERs not only at mRNA level (Supplemental Figure 1), but also at protein level (Fig. 1a), which was the first indication that high HSF1 levels may be associated with the CSC phenotype.
Next, we confirmed these observations in other cancer cell lines that represent all three subtypes of breast cancers, including T47D (ER+), MCF7 (ER+), BT20 (TNBC), BT474 (Her2+), and HCC1954 (Her2+). Fluorescence-activated cell sorting (FACS) has been used to isolate CSC and non-CSC subpopulations based on cell surface marker expression [23–28]. In our work, we used CD44/CD166 double staining and found that CD44high/CD166high subpopulation has significantly greater sphere-forming capacity compared to non-CSC (CD44low/CD166low) counterparts (data not shown). By Western blot, we found that HSF1 expression is much higher in these FACS-enriched CSC subpopulations compared to the non-CSCs in all three subtypes of breast cancer cell lines, further supporting a potential correlation between HSF1 expression and CSCs phenotype (Fig. 1b).
In order to exclude artifacts associated with FACS enrichment of CSCs, we used dual-immunofluorescence (IF) staining to examine co-expression of HSF1 and CSC markers in situ. These IF stains demonstrate that HSF1 is highly expressed in 13–35 % of the cells (HSF1high) in all of the breast cancer cell lines we examined (Fig. 1c, Supplemental Table 3). Importantly, this HSF1high subpopulation co-express higher levels of CSC markers (CD44/CD326) (Fig. 1c). In MDA-MB231 (TNBC) cell line, ~35 % cells express high levels of HSF1 and 90 % of these HSF1high cells co-express CD44. In MCF7 (ER+) cell line, 57 % of the HSF1high cells are CD44 high. In T47D (ER+) and BT474 (Her2+), approximately 56–68 % of the HSF1high cells are also CD326 high (Fig. 1c, Supplemental Table 3).
HSF1 is necessary to maintain CSC phenotype in breast cancer cell lines
Given the promising correlation between high HSF1 expression and CSC marker expression, we sought to determine whether HSF1 plays a functional role in CSC regulation. After examining endogenous HSF1 expression levels in twenty-two commonly used breast cancer cell lines, we selected T47D, MCF7, and BT474 cell lines that have high endogenous HSF1 expression levels to examine the changes in CSC phenotype after HSF1 knockdown (Supplemental Fig. 2).
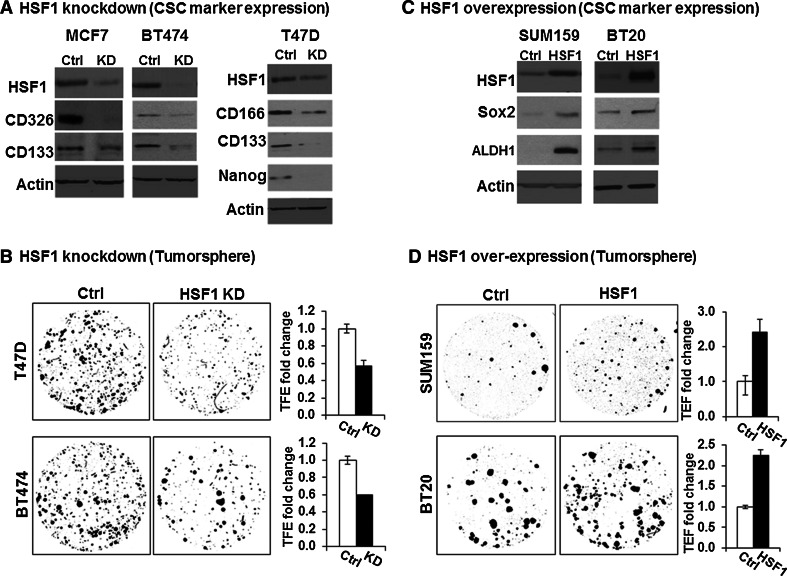
We used the Tet All-in-One inducible lentiviral shRNA knockdown system (Tet-on shHSF1) with non-Target shRNA (NT-shRNA) as control for HSF1 knockdown. The induction efficiency was tested using various doses of DOX and as low as 1 ng/ml was able to achieve HSF1 knockdown (Supplemental Fig. 3). HSF1 expression was reduced after induction of HSF1-shRNA with 1 ng/ml DOX for 48 h (Fig. 2a), which is associated with a decrease in CSC markers CD133/CD326 in MCF7 and BT474, and CD133/CD166/Nanog in T47D [22] (Fig. 2a). Moreover, we found that knockdown of HSF1 significantly decreases 3D tumorsphere formation (Fig. 2b). In control experiments, there was no change in the HSF1 levels, CSC markers, or sphere formation with NT-shRNA cells after DOX stimulation, ensuring the results are specific to HSF1 knockdown (Supplemental Figure 4). There was also no effects on cell viability or proliferation rate in 2D cultures after HSF1 knockdown (data not shown), suggesting that the decrease in sphere formation is not due to a generic suppression of cell proliferation.
Fig. 2.
HSF1 is sufficient and necessary to augment CSC phenotype in multiple cell lines. a HSF1 knockdown decreases CSC-associated marker expression in breast cancer cell lines BT474, T47D, and MCF7. HSF1 knockdown was carried out by adding doxycycline (DOX) at 1 ng/ml for 48 h. The same cell line without DOX addition served as controls. Western blot of protein lysates (30 ug) from HSF1 knockdown (KD) and control cells was performed with HSF1, CD326, CD133, CD166, and Nanog antibodies. β-actin (Actin) was used as loading control. b HSF1 knockdown decreases tumorsphere formation in breast cancer cell lines BT474, T47D, and MCF7. The cells were seeded at 3–5 × 103 cells/well on 6-well low adherent plates, and HSF1 knockdown (KD) was induced by adding DOX 2 days before sphere assay. The same cell line without DOX addition was used as the control (ctrl). The images provide representative examples of spheres that were counted after 10–14 days of incubation under an IF microscope at 2× magnification after DAPI staining using ImageJ. The bar graphs show the average of 3 replicates comparing the tumorsphere formation efficiency (TFE) between HSF1 KD and control. The error bars show standard deviation of the mean of 3 replicates, student t-test (p < 0.05). c HSF1 over-expression increases cancer stem cell (CSC) marker expression in SUM159 and BT20. HSF1 over-expression was carried out by adding doxycycline (DOX) at 1 ng/ml for 48 h. The same cell line without DOX addition was served as control. Western blot of protein lysates (30 ug) from HSF1 over-expression (HSF1) and control cells (Ctrl) was performed with HSF1, Sox2, ALDH1 antibodies. β-actin (Actin) was used as loading control. d HSF1 over-expression increases tumorsphere formation in breast cancer cell lines SUM159 and BT20. The images provided is a representative example of spheres that were counted after 10–14 days under an IF microscope after DAPI staining using ImageJ. The bar graphs show the average of 3 replicates comparing the tumorsphere formation efficiency (TFE) between HSF1 over-expression (HSF1) and control (Ctrl). The error bars show standard deviation of the mean of 3 replicates, student t-test (p < 0.05)
In order to examine the CSC phenotype after HSF1 over-expression, we selected SUM159 and BT20 because of relatively lower expression levels in these lines (Supplemental Fig. 2). The HSF1 over-expression in these two cell lines was achieved using lentiviral tetracycline inducible expression system (TetR-HSF1). The sphere formation assays revealed that HSF1 over-expression significantly increases the CSC markers ALDH1 and SOX2 in both cell lines compared to control group (Ctrl) (Fig. 2c), and induced the number of tumorspheres by >twofold (p < 0.05) (Fig. 2d). This effect was not due to a generic increase in cell proliferation since we did not observe any changes in cell proliferation in routine 2D culture after HSF1 over-expression (data not shown). Taken together, these data suggest that HSF1 is both sufficient and necessary to augment the CSC phenotype.
HSF1 expression confers drug resistance
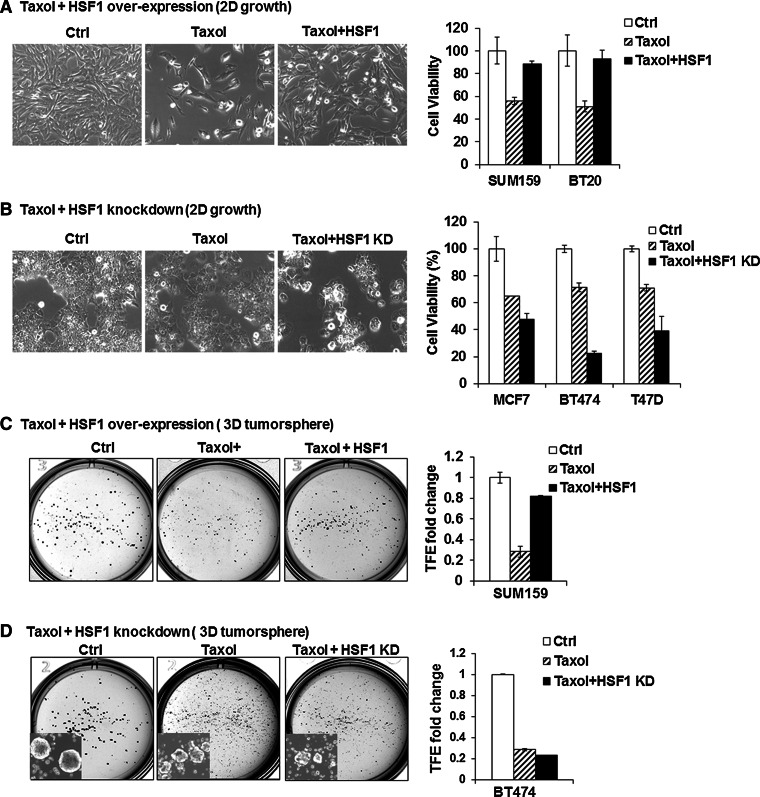
Another feature of CSCs is their relative resistance to chemotherapy [29]. To test whether HSF1 is involved with chemo-resistance, we treated cells with HSF1 over-expression or knockdown with paclitaxel (taxol) and found that HSF1 overexpressing cells are more resistant to taxol treatment compared to control both in BT20 and SUM159 cell lines (Fig. 3a). Conversely, HSF1 knockdown markedly increased cell death after taxol treatment in all three lines tested (BT474, T47D, and MCF7) (Fig. 3b).
Fig. 3.
HSF1 confers drug resistance to breast cancer cells. a HSF1 over-expression increases resistance of cancer cell lines to paclitaxel (Taxol) treatment. HSF1 over-expression was induced with DOX (1 ng/ml) 2 days before drug treatment in SUM159 and BT20. The control cells were treated with vehicle only (Ctrl) or with 2.5 nM Taxol (Taxol) without HSF1 induction with DOX and compared to 2.5 nM Taxol plus HSF1 over-expression (Taxol + HSF1). Cell viability was calculated using Trypan blue exclusion method after 4 days of treatment. The bright field images provide a representative example (10× magnification) and the bar graphs show that the reduction in cell numbers observed with Taxol treatment (striped bars) is prevented with HSF1 over-expression (black bars), compared to control (white bars). Error bars show standard deviation of the mean of 3 replicates (p < 0.05). b HSF1 knockdown increases cancer cell line sensitivity to taxol treatment. HSF1 knockdown was induced with DOX (1 ng/ml) 2 days before drug treatment in MCF7, BT474, and T47D that were treated with vehicle only (Ctrl), 2.5 nM Taxol (Taxol) and 2.5 nM Taxol plus HSF1 knockdown (Taxol + HSF1-KD). Cell viability was calculated using Trypan blue exclusion method after 4 days of treatment. The bright field images provide a representative example (10× magnification) and the bar graphs show that the reduction in cell numbers observed with Taxol treatment (striped bars) is enhanced with HSF1 knockdown (black bars), compared to control (white bars). Error bars show standard deviation of the mean of 3 replicates (p < 0.05). c HSF1 over-expression increases the number of CSCs in breast cancer cell lines treated with taxol. SUM159 cell line expressing inducible HSF1 was plated into T25 flasks with or without DOX (Ctrl or HSF1) for 2 days. Next, the cells were treated with 5 nM taxol for 4 days and allowed to recover in fresh media for 3 days before tumorsphere assays. Tumorsphere formation was carried out by seeding 1 × 104 cells into 6-well low adherent plates. The number of spheres was counted after 10–14 days. Tumorsphere formation efficiency (TFE) was compared between control (Ctrl, vehicle only), Taxol treated (Taxol), and Taxol treated plus HSF1 over-expression (Taxol + HSF1). The bright field images provide a representative example and the bar graphs show that the reduction in sphere numbers observed with Taxol treatment (striped bars) which is prevented with HSF1 over-expression (black bars), compared to control (white bars). Error bars show standard deviation of the mean of 3 replicates (p < 0.05). d HSF1 knockdown inhibits CSCs in breast cancer cells lines treated with taxol. Inducible HSF1 knockdown in BT474 cells was induced by adding DOX for 2 days. Cells were then treated with 5 nM for 4 days. After 3 days of recovery in drug-free medium, tumorsphere formation was carried out as described above and compared between control (Ctrl, vehicle only), taxol treated (Taxol), and Taxol treated plus HSF1 knockdown (Taxol/HSF1 KD) groups. The bright field images provide a representative example and the bar graphs show that the reduction in cell numbers observed with Taxol treatment (striped bars), which is enhanced with HSF1 knockdown (black bars), compared to control (white bars). Error bars show standard deviation of the mean of 3 replicates (p < 0.05). Insets: tumorsphere formation image taken at 4× magnification under bright light field with inverted microscopy
In order to test whether changes in chemotherapy resistance after HSF1 over-expression and knockdown is related to the CSC phenotype, we carried out 3D tumorsphere assays and found that HSF1 over-expression increases sphere formation in SUM159 after taxol treatment (Taxol + HSF1) compared to the taxol alone group (Taxol+) (Fig. 3c). Conversely, in BT474 HSF1 knockdown cells treated with taxol (Taxol + HSF1 KD), there was a small yet statistically significant decrease in number of tumorspheres compared to treatment alone group (Taxol+) (Fig. 3d, insets) [9, 30]. Consistent with our results, Vydra et al. also showed that HSF1 over-expression causes chemo-resistance by increasing side-population (SP) cells in human melanoma [31].
Regulation of CSC phenotype by HSF1
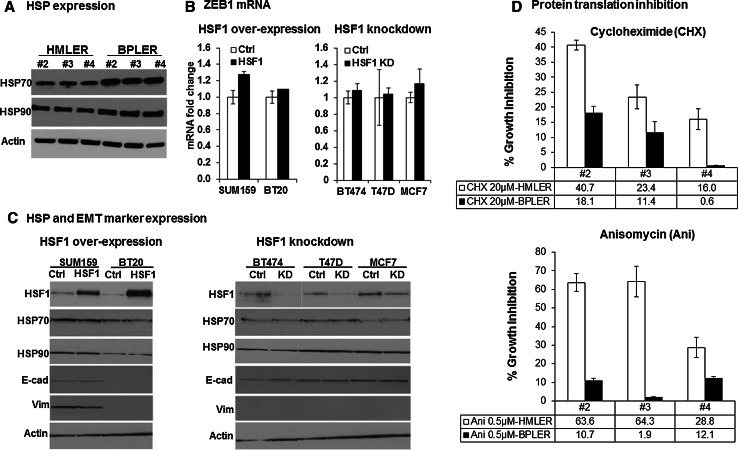
To examine the possible factors that may mediate the actions of HSF1 that effect CSC phenotype, we first analyzed the expression of heat shock proteins in HSF1high CSC-like BPLERs and HSFlow non-CSC-like HMLERs. Compared to the significant differential expression of HSF1 between BPLERs and HMLERs, we only observed very minor changes in the expression of HSP70 and HSP90, two of the main heat shock proteins (Fig. 4a). Next we examined whether HSF1 over-expression increases markers of epithelial-to-mesenchymal transition (EMT), which has been implicated in inducing the CSC phenotype [10]. However, we did not observe any change in EMT markers such as E-cadherin, Vimentin, or ZEB1 with HSF1 over-expression or knockdown (Fig. 4b, c). Likewise, there was no significant change in HSP70 or HSP90 expression after HSF1 knockdown or over-expression in breast cancer cells (Fig. 4c).
Fig. 4.
Mechanisms of CSC regulation by HSF1 a Western blots of three matched pairs of HSF1high CSC-like BPLER vs. HSF1low non-CSC-like HMLER cells (#2-4) reveal only minor differences in heat shock proteins HSP70 and HSP90. The cell lysates were probed with anti-HSP70 and anti-HSP90 antibodies. β-actin (Actin) was used as loading control. b Real-time PCR analysis of ZEB1 mRNA reveals no significant change with HSF1 over-expression (left panel) and knockdown (right panel) after 2 days DOX addition. The Zeb1 mRNA was quantified with real-time PCR after HSF1 over-expression in SUM159 and BT20 cells (black bar, left panel) or knockdown in BT474, T47D, and MCF7 cells (black bar, right panel) compared to control cells without DOX (white bars). GAPDH was used to normalize Ct value. The error bars represent standard deviation of the mean. c HSF1 over-expression or knockdown has no effect on heat shock protein (HSP) or EMT marker expression. Western blot analysis of HSF1, HSP 70, 90, E-cadherin(E-cad), and Vimentin(Vim) after HSF1 over-expression (HSF1) in SUM159 and BT20 cells (left panel) and knockdown (KD) in BT474, T47D, and MCF7 cells (right panel) cells compared to control cells (without DOX) revealed no significant changes. β-actin (Actin) was used as loading control. d HSF1high CSC-like BPLER cells are more sensitive to protein translation inhibition compared to HSF1low non-CSC-like HMLER cells. Three pairs of matched BPLER and HMLER cell lines (#2–4) were treated either with cycloheximide (top panel) or anisomycin (bottom panel) for 8 h, and cultured in drug-free medium for 4 days and cell viability was measured with celltiter-blue assay on day 4. The bar graphs show percent cell viability as compared to control cells (100 %, not shown here). HMLER: white bar; BPLER: black bar
It has been reported that malignant cells upregulate their protein translation to manage the high metabolic stress associated with the malignant phenotype [11]. Santagata et al. found that the increased protein translation in cancer cells may be mediated by HSF1 and it is essential for cancer cell survival [32]. Consistent with this, we found that both knockdown of HSF1 and inhibition of protein translation using Cycloheximide or Anisomycin cause inhibition of HSF1high-CSC-like BPLER proliferation compared to HSF1low non-CSC-like HMLER cell lines (Fig. 4d, Supplemental Figure 5).
Discussion
Here we report that HSF1 plays a role in the regulation of cancer stem cell phenotype in breast cancer cell lines. Previously, we demonstrated that high HSF1 expression is associated with poor prognosis and increased mortality in more than 1800 clinical breast cancer patients’ samples [10]. In this study, we discovered that HSF1 is highly expressed in breast CSC subpopulations. Furthermore, we found that CSC phenotype is augmented by HSF1 over-expression and inhibited by HSF1 knockdown in breast cancer cells lines. Hence, cumulatively, our results suggest that the correlation between high HSF1 expression and poor patient outcome might be partially explained by the actions of HSF1 on CSCs in breast tumor [29, 30, 33, 34].
Together these data suggest that HSF1-mediated augmentation of CSC phenotype involves mechanisms in addition to HSPs and EMT, including HSF1-mediated protein translation that may be feature of CSC survival. Interestingly, a recent genome-wide siRNA screen identified protein degradation and proteasome addiction as a vulnerability of HSF1high CSC-like BPLER cells, which provides independent corroboration of these results [21]. The inhibition of the proteasome reduced growth of established breast cancers in mice and blocked tumor initiation by CSCs and metastasis [21]. It is intriguing that the hypothesis-based studies converge with unbiased high-throughput screens over the protein turn-over machinery.
While the role of HSF1 has been traditionally explored in terms of its role in regulating HSPs and stress response, there is rapidly growing evidence that HSF1 regulates many non-HSPs genes that regulate many cellular processes including protein translation, cell cycle, glucose anabolic metabolism, autophagy, apoptosis, p53, Ras, MAPK, cAMP, PKA, and mTOR pathways [11, 35–37]. Our results suggest that some of these non-HSP targets of HSF1 may be involved in the regulation of CSC phenotype as well, which makes HSF1 as a potential target to develop CSC-specific therapies [32, 38–40].
Electronic supplementary material
Acknowledgments
We thank Dr. Stephen Ethier for providing the SUM159 and SUM190 cell lines. This study was funded by grants from the Florida Bankhead-Coley Cancer Research Program (Grant No: 4CC12), Breast Cancer Research Foundation, NY, 1R01ES024991 and R01CA146445 (to T.A.I.).
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 2.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104(3):973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 4.Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65(20):9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 5.Dean M. Cancer stem cells: implications for cancer causation and therapy resistance. Discov Med. 2005;5(27):278–282. [PubMed] [Google Scholar]
- 6.Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66(12):6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smalley MJ, Dale TC. Wnt signalling in mammalian development and cancer. Cancer Metastasis Rev. 1999;18(2):215–230. doi: 10.1023/A:1006369223282. [DOI] [PubMed] [Google Scholar]
- 8.Dierks C, Beigi R, Guo GR, Zirlik K, Stegert MR, Manley P, Trussell C, Schmitt-Graeff A, Landwerlin K, Veelken H, Warmuth M. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell. 2008;14(3):238–249. doi: 10.1016/j.ccr.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Bhola NE, Balko JM, Dugger TC, Kuba MG, Sanchez V, Sanders M, Stanford J, Cook RS, Arteaga CL. TGF-beta inhibition enhances chemotherapy action against triple-negative breast cancer. J Clin Invest. 2013;123(3):1348–1358. doi: 10.1172/JCI65416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE, Schnitt SJ, Whitesell L, Tamimi RM, Lindquist S, Ince TA. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci USA. 2011;108(45):18378–18383. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, Lindquist S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150(3):549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, Wang S, Liu N, Tang X. Correlation between the expression of Id-1 and hyperthermia-associated molecules in oral squamous cell carcinoma. J Clin Pathol. 2013;66(9):758–763. doi: 10.1136/jclinpath-2011-200287. [DOI] [PubMed] [Google Scholar]
- 13.Scott KL, Nogueira C, Heffernan TP, van Doorn R, Dhakal S, Hanna JA, Min C, Jaskelioff M, Xiao Y, Wu CJ, Cameron LA, Perry SR, Zeid R, Feinberg T, Kim M, Vande Woude G, Granter SR, Bosenberg M, Chu GC, DePinho RA, Rimm DL, Chin L. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell. 2011;20(1):92–103. doi: 10.1016/j.ccr.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang F, Chang R, Yang L. Heat shock factor 1 promotes invasion and metastasis of hepatocellular carcinoma in vitro and in vivo. Cancer. 2012;118(7):1782–1794. doi: 10.1002/cncr.26482. [DOI] [PubMed] [Google Scholar]
- 15.Engerud H, Tangen IL, Berg A, Kusonmano K, Halle MK, Oyan AM, Kalland KH, Stefansson I, Trovik J, Salvesen HB, Krakstad C. High level of HSF1 associates with aggressive endometrial carcinoma and suggests potential for HSP90 inhibitors. Br J Cancer. 2014;111(1):78–84. doi: 10.1038/bjc.2014.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ince TA, Richardson AL, Bell GW, Saitoh M, Godar S, Karnoub AE, Iglehart JD, Weinberg RA. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12(2):160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4(8):e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiederschain D, Wee S, Chen L, Loo A, Yang G, Huang A, Chen Y, Caponigro G, Yao YM, Lengauer C, Sellers WR, Benson JD. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8(3):498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 19.Shaw FL, Harrison H, Spence K, Ablett MP, Simoes BM, Farnie G, Clarke RB. A detailed mammosphere assay protocol for the quantification of breast stem cell activity. J Mammary Gland Biol Neoplas. 2012;17(2):111–117. doi: 10.1007/s10911-012-9255-3. [DOI] [PubMed] [Google Scholar]
- 20.Santagata S, Thakkar A, Ergonul A, Wang B, Woo T, Hu R, Harrell JC, McNamara G, Schwede M, Culhane AC, Kindelberger D, Rodig S, Richardson A, Schnitt SJ, Tamimi RM, Ince TA. Taxonomy of breast cancer based on normal cell phenotype predicts outcome. J Clin Invest. 2014;124(2):859–870. doi: 10.1172/JCI70941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrocca F, Altschuler G, Tan SM, Mendillo ML, Yan H, Jerry DJ, Kung AL, Hide W, Ince TA, Lieberman J. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell. 2013;24(2):182–196. doi: 10.1016/j.ccr.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abigail E. W. C-WL, Tong I. L., et al (2014) Identification of a tumor stem cell-specific function for the histone deacetylases, HDAC1 and HDAC7 in breast and ovarian cancer. submission [DOI] [PMC free article] [PubMed]
- 23.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, Shelton AA, Parmiani G, Castelli C, Clarke MF. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104(24):10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiao J, Hindoyan A, Wang S, Tran LM, Goldstein AS, Lawson D, Chen D, Li Y, Guo C, Zhang B, Fazli L, Gleave M, Witte ON, Garraway IP, Wu H. Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS One. 2012;7(8):e42564. doi: 10.1371/journal.pone.0042564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 26.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci F, Jacquemier J, Xerri L, Dontu G, Stassi G, Xiao Y, Barsky SH, Birnbaum D, Viens P, Wicha MS. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16(1):45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69(4):1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, Wong H, Rosen J, Chang JC. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100(9):672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 31.Vydra N, Toma A, Glowala-Kosinska M, Gogler-Piglowska A, Widlak W. Overexpression of Heat Shock Transcription Factor 1 enhances the resistance of melanoma cells to doxorubicin and paclitaxel. BMC Cancer. 2013;13:504. doi: 10.1186/1471-2407-13-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santagata S, Mendillo ML, Tang YC, Subramanian A, Perley CC, Roche SP, Wong B, Narayan R, Kwon H, Koeva M, Amon A, Golub TR, Porco JA, Jr, Whitesell L, Lindquist S. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science. 2013;341(6143):1238303. doi: 10.1126/science.1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells. 2011;29(1):32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 34.Donovan CA, Pommier RF, Schillace R, O’Neill S, Muller P, Alabran JL, Hansen JE, Murphy JA, Naik AM, Vetto JT, Pommier SJ. Correlation of breast cancer axillary lymph node metastases with stem cell mutations. JAMA Surg. 2013;148(9):873–878. doi: 10.1001/jamasurg.2013.3028. [DOI] [PubMed] [Google Scholar]
- 35.Li D, Yallowitz A, Ozog L, Marchenko N. A gain-of-function mutant p53-HSF1 feed forward circuit governs adaptation of cancer cells to proteotoxic stress. Cell Death Dis. 2014;5:e1194. doi: 10.1038/cddis.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou SD, Prince T, Gong J, Calderwood SK. mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PLoS One. 2012;7(6):e39679. doi: 10.1371/journal.pone.0039679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desai S, Liu Z, Yao J, Patel N, Chen J, Wu Y, Ahn EE, Fodstad O, Tan M. Heat shock factor 1 (HSF1) controls chemoresistance and autophagy through transcriptional regulation of autophagy-related protein 7 (ATG7) J Biol Chem. 2013;288(13):9165–9176. doi: 10.1074/jbc.M112.422071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santagata S, Xu YM, Wijeratne EM, Kontnik R, Rooney C, Perley CC, Kwon H, Clardy J, Kesari S, Whitesell L, Lindquist S, Gunatilaka AA. Using the heat-shock response to discover anticancer compounds that target protein homeostasis. ACS Chem Biol. 2012;7(2):340–349. doi: 10.1021/cb200353m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon YJ, Kim JA, Shin KD, Shin DS, Han YM, Lee YJ, Lee JS, Kwon BM, Han DC. KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. J Biol Chem. 2011;286(3):1737–1747. doi: 10.1074/jbc.M110.179440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Au Q, Zhang Y, Barber JR, Ng SC, Zhang B. Identification of inhibitors of HSF1 functional activity by high-content target-based screening. J Biomol Screen. 2009;14(10):1165–1175. doi: 10.1177/1087057109347472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.