Abstract
The ability of Clostridium thermocellum to rapidly degrade cellulose and ferment resulting hydrolysis products into ethanol makes it a promising platform organism for cellulosic biofuel production via consolidated bioprocessing. Currently, however, ethanol yield is far below theoretical maximum due to branched product pathways that divert carbon and electrons towards formate, H2, lactate, acetate, and secreted amino acids. To redirect carbon and electron flux away from formate, genes encoding pyruvate:formate lyase (pflB) and PFL-activating enzyme (pflA) were deleted. Formate production in the resulting Δpfl strain was eliminated and acetate production decreased by 50 % on both complex and defined medium. The growth rate of the Δpfl strain decreased by 2.9-fold on defined medium and biphasic growth was observed on complex medium. Supplementation of defined medium with 2 mM formate restored Δpfl growth rate to 80 % of the parent strain. The role of pfl in metabolic engineering strategies and C1 metabolism is discussed.
Electronic supplementary material
The online version of this article (doi:10.1007/s10295-015-1644-3) contains supplementary material, which is available to authorized users.
Keywords: Cellulosic ethanol, Clostridium thermocellum, Pyruvate:formate lyase, Metabolic engineering, C1 metabolism
Introduction
Growing global energy demands, rural economic development, the volatile cost of fossil fuels, and environmental concerns have prompted research into the development of sustainable and environmentally benign energy sources. Biofuels provide a promising alternative to petroleum-derived fuels for transportation, one of the largest and fastest growing energy sectors [6]. Currently, bioethanol is a leading candidate that can be used with current technologies as a fuel supplement or replacement. A number of strains that produce ethanol at high yield and titer (e.g., Saccharomyces cerevisiae or Zymomonas mobilis) are available for industrial bioethanol production, but require monosaccharides or disaccharides typically derived from food/feed sources (e.g., corn, beets, sugarcane). These sugars can also be generated through chemical or enzymatic hydrolysis of highly abundant lignocellulosic biomass. However, consolidated bioprocessing (CBP), featuring one-step processing without added enzymes, has potential for lower costs as compared to processes that involve a dedicated step for cellulase production [23–25]. While a number of organisms are capable of cellulase-mediated cellulose hydrolysis and subsequent fermentation to ethanol, none have been yet identified or developed that can produce ethanol at high yields and titer required for commercial production.
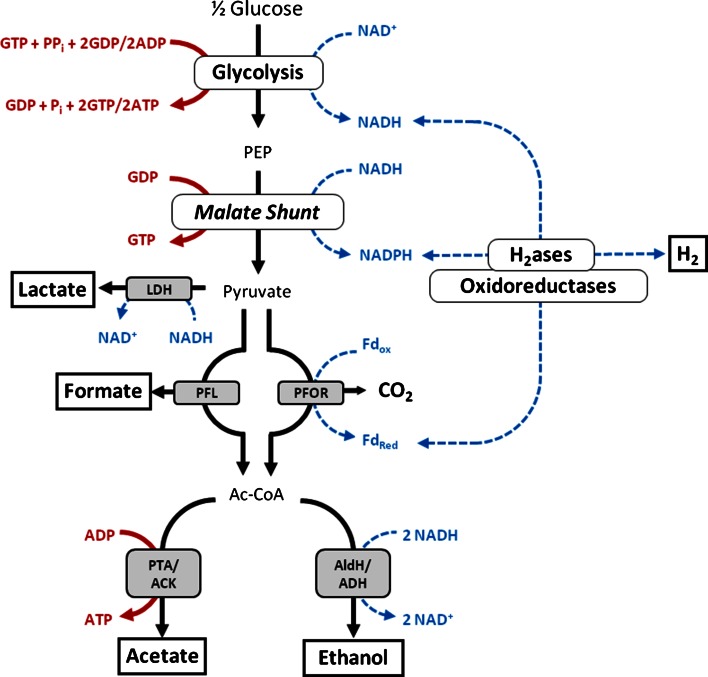
Clostridium thermocellum is a promising candidate for ethanol production via CBP given its inherent ability to rapidly solubilize cellulose and ferment the hydrolysis products to biofuels (i.e., ethanol and H2) [13, 21, 22]. However, branched metabolic pathways divert carbon and/or electrons away from ethanol towards undesired fermentation products including formate, H2, lactate, and acetate (Fig. 1). Many of these pathways have been elucidated through enzymology [15, 20, 28, 32, 37, 38, 44], transcriptomics [5, 29, 42, 43], proteomics [30, 31, 34], and genetics [1, 3, 7, 39]. More recent studies have demonstrated that carbon and electron flux are also diverted towards secreted amino acids [8, 14, 41] and other compounds including pyruvate, malate, fumarate, isobutanol, and butanediol, [12], further limiting ethanol yields.
Fig. 1.
Fermentation pathways in C. thermocellum. Fd ferredoxin; LDH lactate dehydrogenase; PFOR pyruvate: Fd oxidoreductase; PFL pyruvate: formate lyase; PTA/ACK phosphotransacetylase/acetate kinase; AldH/ADH aldehyde dehydrogenase/alcohol dehydrogenase; H 2 ases hydrogenases including ech-type Fd-dependent hydrogenase and bifurcating hydrogenases. Oxidoreductases include NADH dependent reduced ferredoxin: NADP+ oxidoreductase (NfnAB) and NADH:Fd oxidoreductase (RNF)
While a number of studies have demonstrated that manipulation of fermentation conditions can influence product yields [5, 16, 33], engineering of a C. thermocellum strain capable of high-yield ethanol production will require the elimination of the pathways involved in production of competing fermentation products. Recent advances related to genetic tools applicable to C. thermocellum, including electrotransformation protocols [27, 40], positive and negative selection systems used to select for gene deletions [1, 39], and improvement of transformation efficiencies through elimination of Dcm methylation of plasmid DNA [9] have allowed for genetic engineering of C. thermocellum. These tools have been used to begin engineering C. thermocellum to increase ethanol yield. Previous deletion of genes involved in many competing pathways have included phosphotransacetylase (pta; [1, 39]), lactate dehydrogenase (ldh; [1, 2]), malate dehydrogenase (mdh; [7]), malic enzyme (malE; [7]), Ech-type ferredoxin-dependent hydrogenase (ech; [3]), and a [FeFe] hydrogenase maturation factor to inactivate [FeFe] hydrogenases (hydG; [3]). Further, to increase NADH availability for bifunctional aldehyde/alcohol dehydrogenase (adhE), heterologous expression of pyruvate kinase was used to bypass the ‘malate shunt’ to reduce transhydrogenation that generates NADPH from using electrons from NADH [7], and expression of a mutant AdhE allele was also shown to increase ethanol tolerance in C. thermocellum [4].
While most pathways to products other than ethanol have been deleted in C. thermocellum, this is not the case for formate synthesis. Production of reduced compounds such as formate limits the electrons available for ethanol production; therefore, identifying and deleting the gene(s) involved in formate synthesis are important next steps in the engineering of C. thermocellum for high-yield ethanol production for CBP.
In this study, we simultaneously deleted pyruvate:formate lyase (pflB; Clo1313_1717) and Pfl-activating enzyme (pflA; Clo1313_1716) in C. thermocellum in an attempt to increase electron flux towards ethanol and to understand how this mutation impacts growth, end-product synthesis, and amino acid secretion on rich and defined medium.
Materials and methods
Strains, culture conditions, and reagents
Saccharomyces cerevisiae InvSc1 (uracil auxotroph; Life Technologies, Grand Island, NY), used for yeast gap repair cloning, was maintained on YPD medium and grown on SD-ura medium (Sunrise Science Products, San Diego, CA) when selecting for presence of URA3+ plasmids. Escherichia coli Top10 (dam+dcm+; Invitrogen, Carlsbad, CA) and BL21 (dam+dcm−; New England Biolabs, Ipswich, MA) strains were used for plasmid construction and propagated aerobically on LB medium supplemented with 12 μg/ml chloramphenicol as required for plasmid maintenance. Clostridium thermocellum strains were derived from strain DSM 1313 (Deutsche Sammlung von Microorganismen und Zellkulturen, Braunshwieg, Germany) and were routinely grown anaerobically at 55 °C, unless otherwise noted, in a Coy anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) on 5 g/l cellobiose in modified DSM 122 complex medium supplemented with 50 mM MOPS and 10 mM sodium citrate [39] (referred to as CTFUD). Medium was made anaerobic via autoclaving to remove O2 from solution, followed by immediate transfer to the anaerobic chamber to maintain anaerobicity and was supplemented with 10 μg/ml thiamphenicol, 50 μg/ml 5-fluoro-2′-deoxyuridine, or 500 μg/ml 8-azahypoxanthine (Tokyo Chemical Industry, Co., Tokyo, Japan) during C. thermocellum strain construction when appropriate. C. thermocellum strains were grown in either CTFUD or a modified version of chemically defined Medium for Thermophilic Clostridia (MTC; [36]) in which (i) urea concentrations were reduced and medium was supplemented with trace elements and additional vitamins as outlined by [2] and (ii) all stock solutions were filter sterilized rather than autoclaved. This modified version of MTC is now referred to as ‘MTC5’. Tubes containing either CTFUD or MTC5 were made anaerobic by degassing/gassing (2:1 min) ten times with 100 % N2. Final pressure of tubes was equilibrated to 5 psi over ambient pressure. All chemicals were reagent grade and obtained from Sigma-Aldrich (St. Loius, MO) or Fisher Scientific (Waltham, MA) unless otherwise specified.
Plasmid and strain construction
All plasmids used in this study were constructed using yeast gap repair cloning as outlined by Shanks et al. [35]. Plasmid isolation and purification were performed using QIAprep spin miniprep kits (QIAGEN, Germantown, MD). C. thermocellum Δhpt [1] was transformed via electroporation as previously described [9, 27]. The pflAB genes were deleted according to the protocol developed in Argyros et al. [1] and detailed in Olson et al. [27] using plasmid pAMG281 (Accession number KP864661; Online Resource 1). Gene deletion was confirmed using primer sets P1F (5′-GAAATTATACTCCTTATGAAGGCGA-3′) and P1R (5′-TCTGTTCCTTGACTGCTGCAA-3′); P2F (5′-CGGAGCCCAACCTTACAGTAC-3′) and P2R (5′-TATGGAAAGGGTCGGAGTGG-3′); and P3F (5′-ATACTTGATTATTATGAGCGCGG-3′) and P3R (5′-ATTCTCCTGGTTAAGCCTTGTAA-3′) as described below.
Fermentation conditions
Fermentation experiments were carried out in sealed Balch tubes (27 ml; Belco Glass Inc., Vineland, NJ) containing either CTFUD or MTC5 as described above. C. thermocellum inoculum was subcultured a minimum of three times using a 2 % (v/v) inoculum on corresponding medium to prevent carry over. Fermentations were performed on 4.5 g/l cellobiose at 55 °C until all substrate was consumed. Final fermentation products were measured following complete cellobiose utilization (<0.25 mM remaining). Fermentations were performed a minimum of two times with three independent biological replicates each time. For growth curves, cells were grown in 650 μl total volume and growth was monitored spectrophotometrically at OD600 in an Eon Microplate Spectrophotometer (BioTek Instruments Inc., Winooski, VT) situated in a Coy anaerobic chamber.
Analytical methods
Substrate (cellobiose), the cellobiose hydrolysis product glucose, and fermentation products (pyruvate, lactate, acetate, formate, ethanol) were analyzed using a Breeze High Performance Liquid Chromatography system (Waters, Milford, MA) using an Aminex-HPX-87H column (Bio-Rad, Hercules, CA) with a 5 mM sulfuric acid mobile phase. H2 was measured using an Agilent Technologies 6850 Series II Gas Chromatograph (Agilent Technologies, Santa Clara, CA) using a thermoconductivity detector set at 190 °C with an N2 reference flow and a Carbonex 1010 PLOT (30.0 m × 530 μm I.D.; model Supelco 25467) column. Secreted amino acids were measured using an Aracus Amino Acid Analyzer (membraPure, Berlin, Germany) using a T111 Li-cation exchange column with eluents supplied by the manufacturer. Ninhydrin-derived amino acids were measured photometrically at 570 nm with the exception of proline, which was measured at 440 nm. Final pH was measured using an Accument AB15 Basic pH meter (Fisher Scientific; Pittsburg, PA).
Calculations
CO2 produced was calculated based on the expected ratio of C1:C2 compounds and the fact that valine biosynthesis also liberates CO2, whereby CO2 = [(ethanol + acetate) − (formate)] + valine. Ratios of oxidized to reduced fermentation products (O/R) were calculated using reduction values of each fermentation product, calculated as the number of oxygen atoms less one-half the number of hydrogens in each compound [26]. Carbon bound electron equivalents were calculated as described by Harris et al. [10] and were used to determine electron recovery.
Results
Deletion of C. thermocellumpfl eliminates formate and decreases acetate production
The genes encoding pyruvate-formate lyase (pflB) and Pfl-activating enzyme (pflA) were deleted in C. thermocellum Δhpt, confirmed using three primer sets as described in Online Resource 2, and is hereafter referred to as C. thermocellum Δpfl.
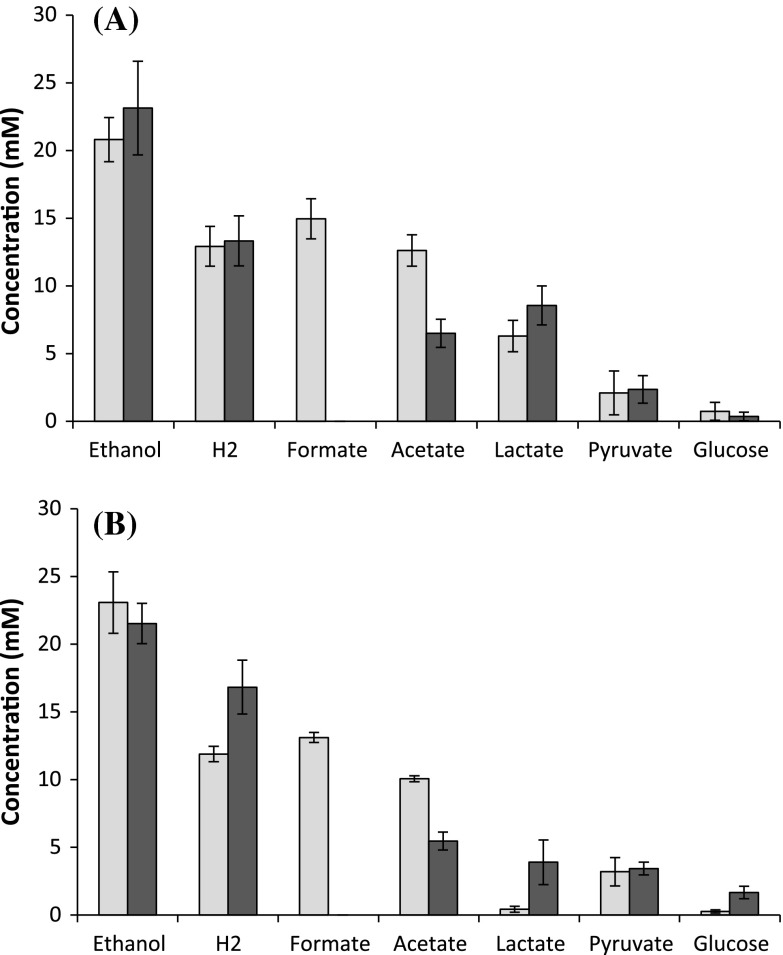
C. thermocellum Δhpt and Δpfl were grown on 4.5 g/l (13.1 mM) cellobiose in complex (CTFUD) medium or defined medium (MTC5). Final fermentation products were measured following nearly complete fermentation of cellobiose (<0.25 mM remaining). Deletion of pfl eliminated formate production and decreased acetate production by ~50 %, regardless of medium used (Fig. 2). However, there were significant medium-dependent differences in other final fermentation products. On CTFUD, no differences were observed in final H2, ethanol, or lactate concentrations in Δpfl cultures when compared to Δhpt. However, when C. thermocellum was grown on MTC5, Δpfl cultures produced 1.4-fold more H2 and 9.3-fold more lactate compared to the wild type along with comparable concentrations of ethanol. Notably, Δhpt cultures grown on CTFUD produced 15-fold more lactate than on MTC5. Residual glucose was detected at the end of growth in all cultures and ranged from 0.3 mM in Δhpt cultures grown on MTC5 to 1.7 mM in Δpfl cultures grown on MTC5 (Fig. 2, Online Resource 3). Secreted pyruvate was also detected on CTFUD (2.1 and 2.4 mM) and MTC5 (3.2 and 3.4 mM) in Δhpt and Δpfl cultures, respectively (Fig. 2, Online Resource 3).
Fig. 2.
Final fermentation products of C. thermocellum strains on a complex medium (CTFUD) and b defined medium (MTC5). All fermentation products were measured upon completion of cellobiose utilization. Light grey bars, Δhpt; Dark grey bars, Δpfl
Given that the addition of sodium bicarbonate to MTC5 rendered accurate measurements of CO2 infeasible, CO2 was calculated based on detected fermentation products (Table 1). As expected, deletion of pfl resulted in an increase in calculated CO2 production of 1.6- and 1.3-fold on CTFUD and MTC5, respectively. Oxidation/reduction balances (O/R) based on formate, ethanol, H2, and calculated CO2 were ~ 1, in support of the validity of our carbon accounting assumptions.
Table 1.
Final product yields and fermentation balances of Δhpt and Δpfl on CTFUD and MTC
| Product yields (mol mol-hexose consumed−1) | O/R balanceb | C recovery (%)c | e− recovery (%)c | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ethanol | H2 | Formate | Acetate | Lactate | COa2 | FPs | AAs | Total | FPs | AAs | Total | ||
| CTFUD | |||||||||||||
| Δhpt | 0.77 | 0.48 | 0.56 | 0.47 | 0.23 | 0.74 | 1.05 | 84.5 | 1.6 | 86.2 | 83.7 | 1.1 | 85.9 |
| Δpfl | 0.87 | 0.50 | 0.00 | 0.25 | 0.32 | 1.19 | 1.09 | 79.3 | −0.7 | 78.5 | 77.5 | −0.1 | 77.5 |
| MTC | |||||||||||||
| Δhpt | 0.87 | 0.45 | 0.50 | 0.38 | 0.02 | 0.83 | 1.04 | 72.5 | 7.7 | 80.2 | 71.8 | 9.0 | 80.8 |
| Δpfl | 0.81 | 0.63 | 0.00 | 0.21 | 0.15 | 1.07 | 1.01 | 73.4 | 3.4 | 76.8 | 73.3 | 4.2 | 77.5 |
Carbon recoveries of fermentation products (FPs), amino acids (AAs), and total secreted products are provided
aCO2 was calculated using C1:C2 ratio using ethanol, acetate and formate concentrations
bO/R balance was determined using formate, calculated CO2 (from C1:C2 ratio), H2, and ethanol
cCarbon and electron recoveries were calculated based on all substrates (cellobiose and glucose) and end-products (ethanol, H2, formate, acetate, lactate, pyruvate) detected, calculated CO2 (from C1:C2 ratio), and secreted amino acids, but exclude biomass measurements
Select amino acids are utilized by C. thermocellum
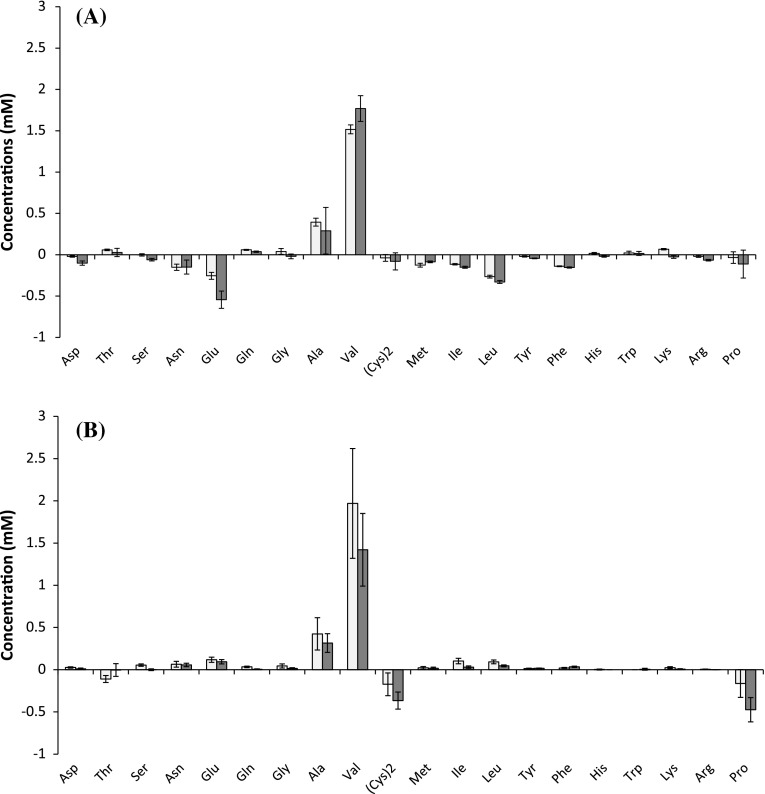
Changes in medium amino acid concentrations at the end of growth are depicted in Fig. 3. Valine and alanine were secreted at the highest levels, regardless of medium or strain used. For CTFUD, which contains yeast extract (Online Resource 3), glutamate and leucine were consumed by both Δhpt (0.4 and 0.3 mM, respectively) and Δpfl (0.7 and 0.3 mM, respectively). All other amino acids were consumed or produced at concentrations less than 0.15 mM (Fig. 3a) on CTFUD. Secreted valine concentrations were 1.5 and 1.8 mM, while secreted alanine concentrations were 0.4 and 0.3 mM, respectively, for Δhpt and Δpfl on CTFUD. While MTC5 was not supplemented with amino acids other than cysteine (used as a reducing agent), 1.4 mM proline and 0.3 mM threonine were detected in uninoculated MTC5. Consumption of both cysteine and proline was observed by both strains in MTC, albeit consumption of each was greater by Δpfl (Fig. 3b). While final concentrations of valine were 2.0 and 1.4 mM for Δhpt and Δpfl, respectively, and those for alanine were 0.4 and 0.3 mM for Δhpt and Δpfl, respectively, concentrations of all other secreted amino acids were less than 0.15 mM.
Fig. 3.
Final secreted amino acid concentrations of C. thermocellum strains on a complex medium (CTFUD) and b defined medium (MTC5). All fermentation products were measured upon completion of cellobiose utilization. Light grey bars Δhpt; Dark grey bars Δpfl; (Cys)2, cystine. Negative values indicate net consumption of the given amino acid. Error bars represent one standard deviation from the mean
Total carbon and electron recoveries in fermentation products were similar between strains under each growth condition and ranged from 77 to 86 % (Table 1). Net production of amino acids accounted for ca. 2 % of final carbon and electron balances in CTFUD-grown Δhpt supernatants, whereas no net difference in amino acids was observed in CTFUD-grown Δpfl supernatants. On MTC5, however, total secreted amino acids accounted for ca. 8 % of carbon and electron recoveries in Δhpt supernatants, and ca. 4 % in Δpfl supernatants (Table 1, Online Resource 3).
Growth rate is altered in C. thermocellum Δpfl
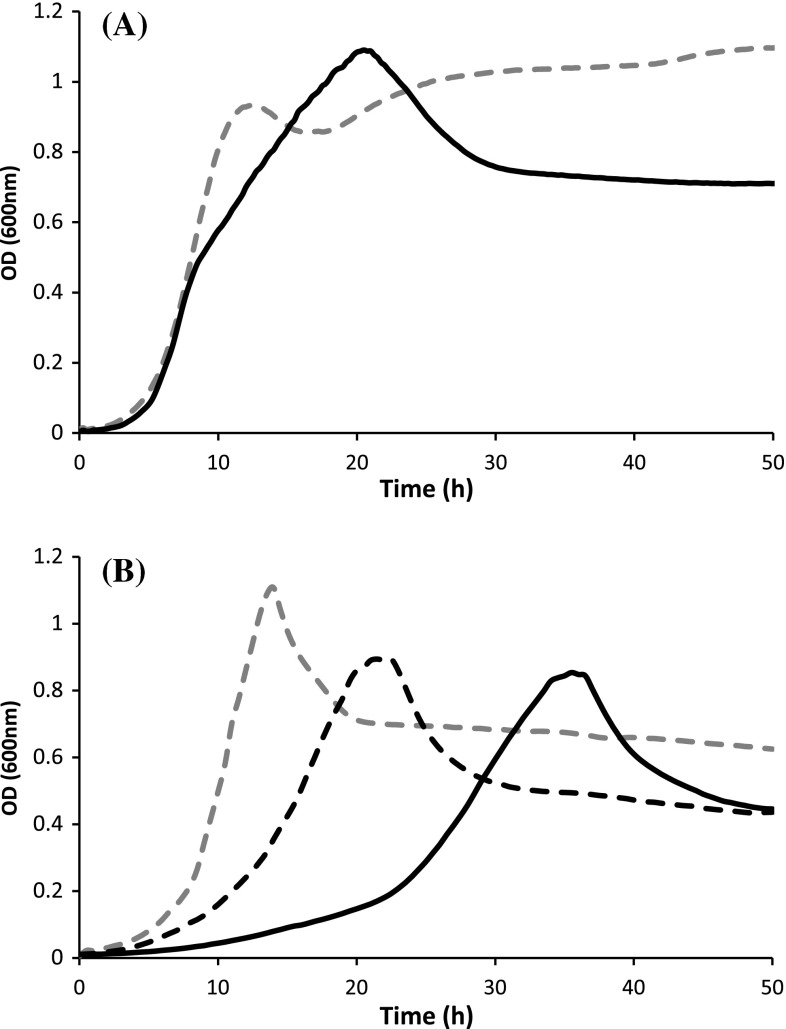
To further examine the effect of pfl deletion in C. thermocellum, we examined growth of the parent strain and the Δpfl mutant on cellobiose in CTFUD and MTC5. Growth of the parent strain and Δpfl were similar on complex CTFUD for the first 7 h, with an exponential-phase generation time of 1.3 h (±0.1) and 1.2 h (±0.2), respectively (Fig. 4a). At an optical density of ca. 0.5, Δpfl began to grow slower [9.3 h (±2.7) doubling time] than the parent strain for an additional 10 h until growth peaked at an OD600 of 1.1. The final pH of Δhpt and Δpfl cultures was 5.9 and 6.3, respectively.
Fig. 4.
Growth profiles of parent and mutant strains on a complex medium (CTFUD) and b defined medium (MTC5). Dashed grey line Δhpt; solid black line Δpfl; dashed black line Δpfl supplemented with 2 mM formate
On defined medium MTC5, both Δhpt and Δpfl exhibited a typical-shaped growth curve (Fig. 4b). While Δhpt grew to the same maximum OD600 (1.1) on defined medium when compared to CTFUD, its exponential-phase generation time increased to 2.0 h (±0.4). Deletion of pfl almost tripled the generation time to 6.0 h (±1.6) and marginally decreased maximum OD600 to 0.9 when compared to the parent strain. The final pH of Δhpt and Δpfl cultures in MTC5 medium was 7.0 and 6.7, respectively. In Staphylococcus aureus, growth of a pfl mutant was improved by addition of 2 mM formate [17]. Therefore, we supplemented MTC5 with 2 mM formate, which decreased the generation time of Δpfl to 3.4 (±0.5) but had negligible impact on maximum OD600.
Discussion
Improvement of C. thermocellum ethanol yields may be achieved through elimination of branched fermentation pathways that divert carbon and electron flux away from ethanol. Previous reports have demonstrated that deletion of genes responsible for acetate [1, 39], lactate [1, 2], and H2 synthesis [3] can increase ethanol yields. Production of formate, which has also been shown to be a major fermentation product of C. thermocellum [37], can also reduce the amount of electrons that are available for ethanol production [31]. Deletion of pfl completely eliminated formate production, demonstrating that it encodes the only functional pathway to formate synthesis in C. thermocellum DSM 1313.
The amount of formate produced by C. thermocellum varied considerably in previous studies and comparisons are complicated by differences in medium composition, pH control, headspace gas accumulation, carbon loading, and batch vs. continuous fermentation. Typical formate yields in C. thermocellum DSM 1313 wild type ranges from 0.05 to 0.15 mol mol-hexose−1 [11, 41, 44] but can be as high as 0.27 mol mol-hexose−1 in Δhpt cultures [2]. In wild type C. thermocellum strain ATCC 27405, reported formate yields range from 0.07 to 0.48 [5, 8, 31, 32]. Here we observed formate yields of 0.57 and 0.50 mol mol-glc−1 for wild-type strains on CTFUD and MTC5, respectively, both of which were on the higher end of the spectrum when compared to previous results. This variability in medium and/or strain-dependent yields could prove problematic for applied processes; however, by deleting pfl, this variable flux to formate is eliminated during C. thermocellum cellulose fermentations.
In the absence of pfl, conversion of pyruvate to acetyl-CoA is only catalyzed by pyruvate:ferredoxin (Fd) oxidoreductase (PFOR), which generates CO2 and reduced Fd (Fig. 1). Calculated final CO2 concentrations were higher in Δpfl when compared to Δhpt by 1.6 and 1.4-fold on CTFUD and MTC5, respectively, suggesting that carbon and electron flux through PFOR is increased. This is in agreement with previous studies in which PFL activity in C. thermocellum ATCC 27405 was inhibited using hypophosphite [31]. Despite the increase of additional reducing equivalents produced in the form of reduced Fd, changes in ethanol production were minimal, whereas acetate production decreased by ca. 50 %, regardless of medium used. Consequently, the increase in calculated CO2 production was only 76 and 49 % of the decrease in formate production on CTFUD and MTC5, respectively, demonstrating that increased flux through PFOR does not fully compensate for loss of pfl, and reduced overall flux from pyruvate to acetyl-CoA may lead to a build-up of intermediates upstream of acetyl-CoA that are diverted away from acetate and ethanol.
On MTC5, deletion of pfl increased carbon and electron flux towards lactate and H2 and decreased carbon flux towards acetate. Given that lactate dehydrogenase requires fructose-1,6-bisphosphate (FbP) as an allosteric activator [28, 32], it may be that higher lactate production in the Δpfl strain is a result of FbP accumulation due to restrictions on the rate of glycolytic flux when conversion of pyruvate to acetyl-CoA is catalyzed by PFOR without PFL. Surprisingly, ethanol production did not increase in the Δpfl strain, consistent with potential limitation by acetyl-CoA or NADH availability. On CTFUD, only a marginal increase in lactate and ethanol was observed, while H2 production did not change, suggesting that carbon and electron flux is diverted elsewhere. The two most abundant secreted amino acids, valine and alanine, are derived from pyruvate; therefore, one might hypothesize that the reduced flux from pyruvate to acetyl-CoA might increase flux to secreted amino acids. However, amino acid secretion in fact decreased in Δpfl cultures when compared to Δhpt, regardless of medium used, suggesting that carbon and electron flux is diverted elsewhere.
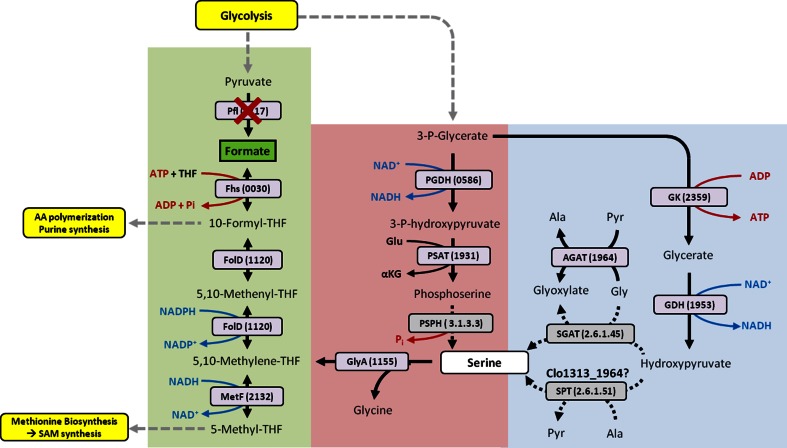
While deletion of pfl eliminated formate production, growth of the Δpfl strain was hindered, especially in minimal medium. On MTC5 the exponential phase growth rate of the Δpfl strain was reduced to 40 % of that of the parent strain, and final OD600 decreased by 21 %. Given that the Δpfl strain produced ~50 % less acetate, a decrease in ATP availability could partially explain lower final growth yields. Interestingly, while deletion of pfl could potentially decrease metabolic flux upstream of acetyl-CoA and, in turn, decrease growth rate, exponential phase growth rate was restored to 80 % of that of the parent strain when MTC5 was supplemented with 2 mM formate. Similar restoration of growth rate was observed with formate supplementation for a Staphylococcus aureus Δpfl mutant [17]. Formate is commonly used for formyl-tetrahydrofolate (THF) synthesis via formate-THF ligase (Clo1313_0030). Formyl-THF in turn acts as a key donor of formyl groups required for purine and formylmethionine synthesis, as well as methyl group donor for synthesis of methionine and S-adenosyl-methionine (Fig. 5). Thus, we hypothesize that elimination of formate production in C. thermocellum has an adverse impact on C1 metabolism, causing a growth defect. Despite this, the Δpfl strain is able to grow in minimal medium without formate supplementation, albeit poorly, indicating that C. thermocellum has an alternate route for synthesizing formyl- and methyl-THF. In the absence of formate supplementation or synthesis via PFL, we postulate that serine may be a precursor for formyl-THF synthesis (Fig. 5). While genomic analysis reveals the presence of all enzymes required for formyl-THF synthesis from serine in C. thermocellum, the pathway for serine biosynthesis is unclear. Possible routes could include phosphoserine phosphatases (PSPH) or serine-pyruvate transaminases (SPT), but none are annotated in C. thermocellum. One possibility is that PSPH activity is present in C. thermocellum but is encoded by an uncharacterized phosphatase. Alternatively, the annotated alanine-glyoxylate transaminase (AGAT) may have SPT activity, synthesizing serine from hydroxypyruvate. Indeed, other studies have demonstrated that the Arabidopsis AGAT can have both AGAT and SPT activities [18, 19]. Regardless of the pathway used to make formyl- and methyl-THF in Δpfl, this pathway is clearly less efficient than the native pathway utilizing formate. Future improvement in formyl- and methyl-THF synthesis in the absence of pfl will likely improve the growth rate of these strains.
Fig. 5.
Proposed formyl-THF synthesis pathway in the presence and absence of formate. Formyl-THF may be synthesized directly from formate via formate-THF ligase (Fhs). In the absence of formate, we propose that serine is a precursor for formyl-THF synthesis. While no annotated phosphoserine phosphatase (PSPH) or serine-pyruvate aminotransferase (SPT) is annotated in C.thermocellum, alanine-glyoxylate transaminase (AGAT) may function as a SPT. Annotated gene numbers (Clo1313) are indicated in brackets. Enzyme E.C. numbers are provided for enzymes which are not annotated in C. thermocellum. THF tetrahydrofolate; GK glycerate kinase; GDH glycerate dehydrogenase; PGDH 3-phosphoglycerate dehydrogenase; PSAT phosphoserine aminotransferase); SGAT serine-glyoxylate aminotransferase; GlyA serine hydroxymethyltransferase; MetF methylene-THF reductase; FolD bifunctional methylene-THF dehydrogenase/methylene-THF cyclohydrolase; Pfl pyruvate:formate lyase
Conclusion
In this study, we have eliminated formate production in C. thermocellum by deleting genes encoding PFL and PFL-activating enzyme. Redirecting metabolic flux away from PFL towards PFOR could have generated additional electrons available for ethanol production. Although ethanol yields did not increase, other fermentation products that act as electron sinks (i.e., H2 and lactate) did increase. Thus, future strategies to improve ethanol yields may involve deletion of lactate and H2-formation pathways in conjunction with deletion of PFL to further limit electron flux towards ethanol. Here we demonstrate that formate availability is important in maintaining growth rate in C. thermocellum and propose that it acts as a precursor for methyl- and formyl-THF biosynthesis, and ultimately methionine, purine and formylmethionine synthesis. Thus, future improvement in formyl-THF synthesis in the absence of pfl will likely improve the growth rate of these strains, which will be required to reduce energy inputs and costs of large-scale fermentations.
Electronic supplementary material
Supplementary material 1 (DOCX 44 kb) Online Resource 1 Key elements of pAMG281 used for deletion of PFL. CEN6/ARSH4 origin, yeast origin of replication; URA3, orotidine 5′-phosphate decarboxylase; bla, beta-lactamase; pNW33 N origin, C. thermocellum origin of replication; P-cbp, C. thermocellum cellobiose phosphorylase promoter; tdk, thymidine kinase; pUC origin, E. coli origin of replication; CYC1 term, Saccharomyces cerevisiae cytochrome C1 transcriptional terminator; pfl up, upstream sequence of homology to pflA; pfl down, downstream sequence of homology to pflB; P-gapD, C. thermocellum glyceraldehyde-3-phosphate dehydrogenase promoter; cat, chloramphenicol acetyltransferase; hpt, hypoxanthine phosphoribosyltransferase; t1t2, T1-T2 terminator; pfl internal, internal sequence of homology to pflB. Full plasmid sequence can be obtained by accessing GenBank accession number KP864661
Supplementary material 2 (DOCX 385 kb) Online Resource 2 Deletion of pfl and pfl-AE; overview and confirmation. A) The pfl and pfl-AE locus was deleted according to the protocol outlined by Argyrose et al. (2011). Primer binding sites used to amplify pfl fragments (P1 and P2) and the locus encompassing pfl and pfl-AE (P3), and corresponding expected product sizes in the parent and mutant strain are indicated. B) PCR confirmation of pfl and pfl-AE deletion. Primer sets P1 and P2 amplified 705 bp and 707 bp fragments of the pfl gene, respectively, in the Δhpt strain, but not in Δpfl. Primer set P3 amplified the chromosomal region that includes pfl, and results in a 5116 bp fragment in the parent strain and a 2105 bp fragment in Δpfl, confirming deletion of pfl. Absence of P1 and P2 amplicons and a reduction in size of the P3 amplicon confirm deletion of pfl and pfl-AE in the mutant strain
Supplementary material 3 (XLSX 50 kb) Online Resource 3 Fermentation product, amino acid, and growth data with corresponding product yields and O/R, carbon, and electron balances
Acknowledgments
This work was supported by the BioEnergy Science Center, U.S. DOE Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. DOE under contract DE-AC05-00OR22725. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Compliance with ethical standards
Conflict of interest
Lee Lynd is an employee of Mascoma Corporation, which has a financial interest in C. thermocellum
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
References
- 1.Argyros DA, Tripathi SA, Barrett TF, Rogers SR, Feinberg LF, Olson DG, Foden JM, Miller BB, Lynd LR, Hogsett DA, Caiazza NC. High ethanol titers from cellulose using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol. 2011;77:8288–8294. doi: 10.1128/AEM.00646-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas R, Prabhu S, Lynd LR, Guss AM. Increase in Ethanol Yield via Elimination of Lactate Production in an Ethanol-Tolerant Mutant of Clostridium thermocellum. PLoS One. 2014;9:e86389. doi: 10.1371/journal.pone.0086389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas R, Zheng T, Olson DG, Lynd LR, Guss AM (2015) Elimination of hydrogenase active site assembly blocks H2 production and increases ethanol yield in Clostridium thermocellum. Biotechnol Biofuels 8. doi:10.1186/s13068-015-0204-4 [DOI] [PMC free article] [PubMed]
- 4.Brown SD, Guss AM, Karpinets TV, Parks JM, Smolin N, Yang S, Land ML, Klingeman DM, Bhandiwad A, Rodriguez M, Jr, Raman B, Shao X, Mielenz JR, Smith JC, Keller M, Lynd LR. Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc Natl Acad Sci U S A. 2011;108:13752–13757. doi: 10.1073/pnas.1102444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carere CR, Rydzak T, Cicek N, Levin DB, Sparling R. Role of transcription and enzyme activities in redistribution of carbon and electron flux in response to N(2) and H(2) sparging of open-batch cultures of Clostridium thermocellum ATCC 27405. Appl Microbiol Biotechnol. 2014;98:2829–2840. doi: 10.1007/s00253-013-5500-y. [DOI] [PubMed] [Google Scholar]
- 6.Chu S, Goldemburg J. Lighting the way: toward a sustainable energy future. Amsterdam: InterAcademy Council; 2007. [Google Scholar]
- 7.Deng Y, Olson DG, Zhou J, Herring CD, Joe Shaw A, Lynd LR. Redirecting carbon flux through exogenous pyruvate kinase to achieve high ethanol yields in Clostridium thermocellum. Metab Eng. 2013;15:151–158. doi: 10.1016/j.ymben.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Ellis LD, Holwerda EK, Hogsett D, Rogers S, Shao X, Tschaplinski T, Thorne P, Lynd LR. Closing the carbon balance for fermentation by Clostridium thermocellum (ATCC 27405) Bioresour Technol. 2012;103:293–299. doi: 10.1016/j.biortech.2011.09.128. [DOI] [PubMed] [Google Scholar]
- 9.Guss AM, Olson DG, Caiazza NC, Lynd LR. Dcm methylation is detrimental to plasmid transformation in Clostridium thermocellum. Biotechnol Biofuels. 2012;5:30. doi: 10.1186/1754-6834-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris RF, Adams SS. Determination of the carbon-bound electron composition of microbial cells and metabolites by dichromate oxidation. Appl Environ Microbiol. 1979;37:237–243. doi: 10.1128/aem.37.2.237-243.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holwerda EK, Hirst KD, Lynd LR. A defined growth medium with very low background carbon for culturing Clostridium thermocellum. J Ind Microbiol Biotechnol. 2012;39:943–947. doi: 10.1007/s10295-012-1091-3. [DOI] [PubMed] [Google Scholar]
- 12.Holwerda EK, Thorne PG, Olson DG, Amador-Noguez D, Engle NL, Tschaplinski TJ, van Dijken JP, Lynd LR. The exometabolome of Clostridium thermocellum reveals overflow metabolism at high cellulose loading. Biotechnol Biofuels. 2014;7:155. doi: 10.1186/s13068-014-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Islam R, Cicek N, Sparling R, Levin D. Influence of initial cellulose concentration on the carbon flow distribution during batch fermentation by Clostridium thermocellum ATCC 27405. Appl Microbiol Biotechnol. 2009;82:141–148. doi: 10.1007/s00253-008-1763-0. [DOI] [PubMed] [Google Scholar]
- 14.Kridelbaugh DM, Nelson J, Engle NL, Tschaplinski TJ, Graham DE. Nitrogen and sulfur requirements for Clostridium thermocellum and Caldicellulosiruptor bescii on cellulosic substrates in minimal nutrient media. Bioresour Technol. 2013;130:125–135. doi: 10.1016/j.biortech.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Lamed R, Zeikus JG. Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii. J Bacteriol. 1980;144:569–578. doi: 10.1128/jb.144.2.569-578.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamed RJ, Lobos JH, Su TM. Effects of stirring and hydrogen on fermentation products of Clostridium thermocellum. Appl Environ Microbiol. 1988;54:1216–1221. doi: 10.1128/aem.54.5.1216-1221.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leibig M, Liebeke M, Mader D, Lalk M, Peschel A, Gotz F. Pyruvate formate lyase acts as a formate supplier for metabolic processes during anaerobiosis in Staphylococcus aureus. J Bacteriol. 2011;193:952–962. doi: 10.1128/JB.01161-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liepman AH, Olsen LJ. Peroxisomal alanine: glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. Plant J. 2001;25:487–498. doi: 10.1046/j.1365-313x.2001.00961.x. [DOI] [PubMed] [Google Scholar]
- 19.Liepman AH, Olsen LJ. Alanine aminotransferase homologs catalyze the glutamate:glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis. Plant Physiol. 2003;131:215–227. doi: 10.1104/pp.011460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin WR, Peng Y, Lew S, Lee CC, Hsu JJ, Hamel JF, Demain AL. Purification and characterization of acetate kinase from Clostridium thermocellum. Tetrahedron. 1998;54:15915–15925. doi: 10.1016/S0040-4020(98)01001-1. [DOI] [Google Scholar]
- 21.Lynd LR, Grethlein HE. Hydrolysis of dilute acid pretreated mixed hardwood and purified microcrystalline cellulose by cell-free broth from Clostridium thermocellum. Biotechnol Bioeng. 1987;29:92–100. doi: 10.1002/bit.260290114. [DOI] [PubMed] [Google Scholar]
- 22.Lynd LR, Grethlein HE, Wolkin RH. Fermentation of cellulosic substrates in batch and continuous culture by Clostridium thermocellum. Appl Environ Microbiol. 1989;55:3131–3139. doi: 10.1128/aem.55.12.3131-3139.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynd LR, Laser MS, Bransby D, Dale BE, Davison B, Hamilton R, Himmel M, Keller M, McMillan JD, Sheehan J, Wyman CE. How biotech can transform biofuels. Nat Biotechnol. 2008;26:169–172. doi: 10.1038/nbt0208-169. [DOI] [PubMed] [Google Scholar]
- 24.Lynd LR, van Zyl WH, McBride JE, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16:577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol R. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moat AG, Foster JW, Spector MP. Microbial Physiology. 4. New York: Wiley-Liss Inc.; 2002. [Google Scholar]
- 27.Olson DG, Lynd LR. Transformation of Clostridium thermocellum by electroporation. Method Enzymol. 2012;510:317–330. doi: 10.1016/B978-0-12-415931-0.00017-3. [DOI] [PubMed] [Google Scholar]
- 28.Ozkan M, Yilmaz EI, Lynd LR, Ozcengiz G. Cloning and expression of the Clostridium thermocellum L-lactate dehydrogenase gene in Escherichia coli and enzyme characterization. Can J Microbiol. 2004;50:845–851. doi: 10.1139/w04-071. [DOI] [PubMed] [Google Scholar]
- 29.Raman B, McKeown CK, Rodriguez M, Jr, Brown SD, Mielenz JR. Transcriptomic analysis of Clostridium thermocellum ATCC 27405 cellulose fermentation. BMC Microbiol. 2011;11:134. doi: 10.1186/1471-2180-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raman B, Pan C, Hurst GB, Rodriguez M, Jr, McKeown CK, Lankford PK, Samatova NF, Mielenz JR. Impact of pretreated Switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis. PLoS One. 2009;4:e5271. doi: 10.1371/journal.pone.0005271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rydzak T, Grigoryan M, Cunningham ZJ, Krokhin OV, Ezzati P, Cicek N, Levin DB, Wilkins JA, Sparling R. Insights into electron flux through manipulation of fermentation conditions and assessment of protein expression profiles in Clostridium thermocellum. Appl Microbiol Biotechnol. 2014;98:6497–6510. doi: 10.1007/s00253-014-5798-0. [DOI] [PubMed] [Google Scholar]
- 32.Rydzak T, Levin DB, Cicek N, Sparling R. Growth phase-dependant enzyme profile of pyruvate catabolism and end-product formation in Clostridium thermocellum ATCC 27405. J Biotechnol. 2009;140:169–175. doi: 10.1016/j.jbiotec.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 33.Rydzak T, Levin DB, Cicek N, Sparling R. End-product induced metabolic shifts in Clostridium thermocellum ATCC 27405. Appl Microbiol Biot. 2011;92:199–209. doi: 10.1007/s00253-011-3511-0. [DOI] [PubMed] [Google Scholar]
- 34.Rydzak T, McQueen PD, Krokhin OV, Spicer V, Ezzati P, Dwivedi RC, Shamshurin D, Levin DB, Wilkins JA, Sparling R. Proteomic analysis of Clostridium thermocellum core metabolism: relative protein expression profiles and growth phase-dependent changes in protein expression. BMC Microbiol. 2012;12:214. doi: 10.1186/1471-2180-12-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shanks RMQ, Kadouri DE, MacEachran DP, O’Toole GA. New yeast recombineering tools for bacteria. Plasmid. 2009;62:88–97. doi: 10.1016/j.plasmid.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao X, Raman B, Zhu M, Mielenz JR, Brown SD, Guss AM, Lynd LR. Mutant selection and phenotypic and genetic characterization of ethanol-tolerant strains of Clostridium thermocellum. Appl Microbiol Biot. 2011;92:641–652. doi: 10.1007/s00253-011-3492-z. [DOI] [PubMed] [Google Scholar]
- 37.Sparling R, Islam R, Cicek N, Carere C, Chow H, Levin DB. Formate synthesis by Clostridium thermocellum during anaerobic fermentation. Can J Microbiol. 2006;52:681–688. doi: 10.1139/w06-021. [DOI] [PubMed] [Google Scholar]
- 38.Taillefer M, Rydzak T, Levin DB, Oresnik IJ, Sparling R. Reassessment of the Transhydrogenase ‘Malate Shunt’ in Clostridium thermocellum ATCC 27405 through kinetic characterization of malic enzyme and malate dehydrogenase. Appl Environ Microbiol. 2015 doi: 10.1128/AEM.03360-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tripathi SA, Olson DG, Argyros DA, Miller BB, Barrett TF, Murphy DM, McCool JD, Warner AK, Rajgarhia VB, Lynd LR, Hogsett DA, Caiazza NC. Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl Environ Microbiol. 2010;76:6591–6599. doi: 10.1128/AEM.01484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyurin MV, Desai SG, Lynd LR. Electrotransformation of Clostridium thermocellum. Appl Environ Microbiol. 2004;70:883–890. doi: 10.1128/AEM.70.2.883-890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Veen D, Lo J, Brown SD, Johnson CM, Tschaplinski TJ, Martin M, Engle NL, van den Berg RA, Argyros AD, Caiazza NC, Guss AM, Lynd LR. Characterization of Clostridium thermocellum strains with disrupted fermentation end-product pathways. J Ind Microbiol Biot. 2013;40:725–734. doi: 10.1007/s10295-013-1275-5. [DOI] [PubMed] [Google Scholar]
- 42.Wilson CM, Yang S, Rodriguez M, Jr, Ma Q, Johnson CM, Dice L, Xu Y, Brown SD. Clostridium thermocellum transcriptomic profiles after exposure to furfural or heat stress. Biotechnol Biofuels. 2013;6:131. doi: 10.1186/1754-6834-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang S, Giannone RJ, Dice L, Yang ZK, Engle NL, Tschaplinski TJ, Hettich RL, Brown SD. Clostridium thermocellum ATCC27405 transcriptomic, metabolomic and proteomic profiles after ethanol stress. BMC Genom. 2012;13:336. doi: 10.1186/1471-2164-13-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou J, Olson DG, Argyros DA, Deng Y, van Gulik WM, van Dijken JP, Lynd LR. Atypical glycolysis in Clostridium thermocellum. Appl Environ Microbiol. 2013;79:3000–3008. doi: 10.1128/AEM.04037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (DOCX 44 kb) Online Resource 1 Key elements of pAMG281 used for deletion of PFL. CEN6/ARSH4 origin, yeast origin of replication; URA3, orotidine 5′-phosphate decarboxylase; bla, beta-lactamase; pNW33 N origin, C. thermocellum origin of replication; P-cbp, C. thermocellum cellobiose phosphorylase promoter; tdk, thymidine kinase; pUC origin, E. coli origin of replication; CYC1 term, Saccharomyces cerevisiae cytochrome C1 transcriptional terminator; pfl up, upstream sequence of homology to pflA; pfl down, downstream sequence of homology to pflB; P-gapD, C. thermocellum glyceraldehyde-3-phosphate dehydrogenase promoter; cat, chloramphenicol acetyltransferase; hpt, hypoxanthine phosphoribosyltransferase; t1t2, T1-T2 terminator; pfl internal, internal sequence of homology to pflB. Full plasmid sequence can be obtained by accessing GenBank accession number KP864661
Supplementary material 2 (DOCX 385 kb) Online Resource 2 Deletion of pfl and pfl-AE; overview and confirmation. A) The pfl and pfl-AE locus was deleted according to the protocol outlined by Argyrose et al. (2011). Primer binding sites used to amplify pfl fragments (P1 and P2) and the locus encompassing pfl and pfl-AE (P3), and corresponding expected product sizes in the parent and mutant strain are indicated. B) PCR confirmation of pfl and pfl-AE deletion. Primer sets P1 and P2 amplified 705 bp and 707 bp fragments of the pfl gene, respectively, in the Δhpt strain, but not in Δpfl. Primer set P3 amplified the chromosomal region that includes pfl, and results in a 5116 bp fragment in the parent strain and a 2105 bp fragment in Δpfl, confirming deletion of pfl. Absence of P1 and P2 amplicons and a reduction in size of the P3 amplicon confirm deletion of pfl and pfl-AE in the mutant strain
Supplementary material 3 (XLSX 50 kb) Online Resource 3 Fermentation product, amino acid, and growth data with corresponding product yields and O/R, carbon, and electron balances