Abstract
Centromeres of higher eukaryotes are epigenetically defined by the centromere specific histone H3 variant CENP-ACID. CENP-ACID builds the foundation for the assembly of a large network of proteins. In contrast to mammalian systems, the protein composition of Drosophila centromeres has not been comprehensively investigated. Here we describe the proteome of Drosophila melanogaster centromeres as analyzed by quantitative affinity purification-mass spectrometry (AP-MS). The AP-MS input chromatin material was prepared from D. melanogaster cell lines expressing CENP-ACID or H3.3 fused to EGFP as baits. Centromere chromatin enriched proteins were identified based on their relative abundance in CENP-ACID–GFP compared to H3.3-GFP or mock affinity-purifications. The analysis yielded 86 proteins specifically enriched in centromere chromatin preparations.
The data accompanying the manuscript on this approach (Barth et al., 2015, Proteomics 14:2167-78, DOI: 10.1002/pmic.201400052) has been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD000758.
Keywords: CENP-A, CID, Centromere, Drosophila, Chromatin proteomics
|
Specifications Table | |
| Subject area | Biology |
| More specific subject area | Chromatin research, Drosophila centromere composition |
| Type of data | Figures, Table |
| How data was acquired | Mass spectrometry. LC–MS/MS (LC: Ultimate 3000 HPLC system, Thermo-Fisher Scientific. MS: Orbitrap XL, Thermo-Fisher Scientific) |
| Data format | Raw (*.raw), MaxQuant output text files (*.zip) and search parameters (*.xml) |
| Experimental factors | Micrococcal nuclease solubilized chromatin from GFP-fusion H3 variant expressing D. melanogaster cell lines analyzed by affinity purification-mass spectrometry. |
| Experimental features | Micrococcal nuclease solubilization of chromatin, anti-GFP affinity purification, SDS-PAGE, in-gel tryptic digestion, LC–MS/MS analysis of peptides, quantitative comparison of protein enrichment in CENP-ACID vs. H3.3 vs. mock AP-MS samples |
| Data source location | Munich, Germany |
| Data accessibility | Uploaded to the ProteomeXchange Consortium webpage. Dataset identifier PXD000758. |
| http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD000758 | |
Value of the data
-
•
Analysis of the Drosophila centromere proteome
-
•
Identification of uncharacterized potential centromere components in Drosophila
-
•
Quantitative analysis of the composition of specific chromatin regions
1. Experimental design and data
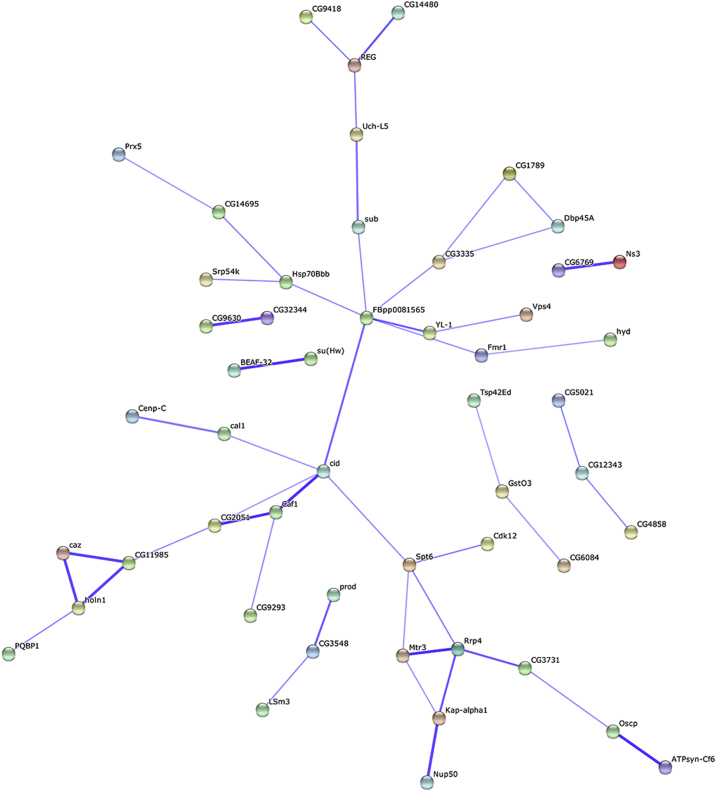
Fig. 1 illustrates the general workflow of the AP-MS approach (adapted from [1]). In order to discriminate centromere specific proteins from proteins abundantly found in chromatin, we quantitatively compared proteomes isolated from different chromatin regions. To biochemically isolate these different chromatin regions, we made use Drosophila cell lines expressing GFP-tagged histone H3 variants as baits. These were either the replacement variant H3.3, which is enriched in euchromatin, or the centromere specific H3 variant CENP-ACID. Chromatin from these cells and the parental cell line was isolated and solubilized by Micrococcal Nuclease digestion. This soluble extract served as input material for anti GFP-affinity purification, thereby enriching chromatin fragments together with the associated proteins. Repeated washes were performed to remove unspecifically bound contaminants. The associated proteins were eluted under denaturating conditions and fractionated via SDS-PAGE. After in gel-tryptic digestion, peptides were extracted, concentrated and analyzed by LC–MS/MS. Three independent AP-MS experiments were performed per cell line used. Intensity-based absolute quantification (iBAQ) values from the output of the MaxQuant software package were used as a measure for the abundance of identified proteins. Average iBAQ values were calculated for the different samples and in case the protein was not detected, the iBAQ values were imputed from a random distribution (see Section 2.5). Centromere enrichment was calculated by dividing the average iBAQ value for each protein in the CENP-ACID–GFP purification by the corresponding iBAQ value in the chromatin purification from untransfected parental or H3.3-GFP expressing cell lines, respectively. A factor was considered centromere specific if its log2-fold enrichment over both controls was more than four. Using these criteria, we identified 86 proteins that were specifically enriched in CENP-ACID–GFP containing chromatin (Table 1 and [1]). Known CENP-ACID-binding proteins such as Cal1, the centromeric proteins CENP-C, or CAF-1 were also found enriched in CENP-ACID chromatin, demonstrating the general applicability of the technique to detect proteins enriched in centromeric chromatin [2]. While centromere association of most of the 86 identified proteins has not been reported so far, several associations among the proteins are predicted using the “Search Tool for the Retrieval of Interacting Genes/Proteins” (STRING) (Fig. 2) [3]. This indicates that a complex network of interactions contributes to centromere function or maintenance.
Fig. 1.
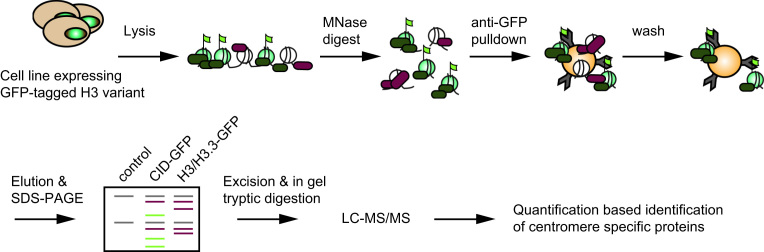
Scheme of experimental strategy. Chromatin of Drosophila control cells or cells stably expressing CENP-ACID–GFP or H3.3-GFP was digested with MNase and anti-GFP affinity purification was performed on the digested soluble chromatin. After washing steps, proteins were eluted by boiling in Laemmli buffer and separated by SDS-PAGE. Following excision from the gel and in gel tryptic digestion, peptide samples were analyzed by LC–MS/MS. (Adapted from [1].)
Table 1.
86 Proteins specifically associate with centromere chromatin List of 86 CENP-ACID chromatin co-purifying proteins ranked according to their calculated enrichment over mock purification.
| Name | FBpp | FBgn | Uniprot | CID-GFP/control | CID-GFP/H3.3-GFP | ||
|---|---|---|---|---|---|---|---|
| log2 | P-value | log2 | P-value | ||||
| cid | FBpp0086787 | FBgn0040477 | Q9V6Q2 | 15.9±1.1E−01 | 1.60E−02 | 11.1±4.5E−05 | 5.80E−02 |
| CG14480 | FBpp0086006 | FBgn0034242 | Q7JWU9 | 11.8±8.7E−02 | 1.70E−05 | 11.6±7.4E−02 | 1.40E−05 |
| cal1 | FBpp0082841 | FBgn0038478 | Q9VEN2 | 11.3±9.1E−02 | 1.60E−05 | 11.2±7.6E−02 | 1.70E−05 |
| CG2051 | FBpp0078319 | FBgn0037376 | Q0KIB3 | 10.8±1.0E−05 | 5.40E−02 | 8.3±3.1E−07 | 1.10E−01 |
| CG13117 | FBpp0079460 | FBgn0032140 | Q9VL93 | 9.2±1.1E−01 | 7.60E−04 | 9.6±2.6E−02 | 6.50E−04 |
| CG34191 | FBpp0111299 | FBgn0085220 | A8DYH2 | 9.2±1.8E−01 | 3.50E−04 | 9.5±1.3E−01 | 3.00E−04 |
| CG6769 | FBpp0074267 | FBgn0030878 | Q9VX08 | 8.6±9.2E−02 | 2.20E−04 | 8.4±7.2E−02 | 2.60E−04 |
| Vps4 | FBpp0074278 | FBgn0027605 | Q9Y162 | 8.0±4.3E−01 | 9.20E−02 | 7.5±4.0E−01 | 1.00E−01 |
| CG4972 | FBpp0079578 | FBgn0032217 | Q9VKZ7 | 7.7±1.6E−01 | 6.20E−05 | 7.6±4.7E−02 | 5.30E−05 |
| Caf1 | FBpp0291059 | FBgn0263979 | E1JIL4 | 7.6±8.6E−03 | 1.90E−04 | 7.5±1.1E−02 | 1.70E−04 |
| LSm3 | FBpp0083893 | FBgn0051184 | Q8IMX8 | 7.5±7.6E−02 | 3.90E−04 | 7.7±5.4E−02 | 2.50E−04 |
| Kap-alpha1 | FBpp0074650 | FBgn0024889 | O76521 | 7.5±4.4E−01 | 1.10E−02 | 8.3±2.5E−01 | 4.60E−03 |
| REG | FBpp0073561 | FBgn0029133 | Q9V3P3 | 7.3±1.1E−04 | 4.00E−01 | 13.4±3.8E−02 | 6.40E−02 |
| ifc | FBpp0078810 | FBgn0001941 | Q94515 | 7.3±9.8E−02 | 2.20E−04 | 7.1±6.8E−02 | 2.60E−04 |
| CG11985 | FBpp0081484 | FBgn0040534 | Q9VHI4 | 7.2±7.5E−02 | 7.10E−04 | 7.5±6.7E−02 | 3.50E−04 |
| CG9293 | FBpp0080103 | FBgn0032516 | Q9VJY8 | 7.1±1.7E−01 | 3.70E−05 | 7.1±6.2E−02 | 4.30E−05 |
| CG8891 | FBpp0078684 | FBgn0031663 | Q9VMW7 | 7.1±1.2E−01 | 7.70E−03 | 7.4±1.2E−01 | 5.00E−03 |
| CG3548 | FBpp0072222 | FBgn0035033 | Q9W140 | 7.0±1.0E−01 | 2.00E−04 | 7.0±4.9E−02 | 2.50E−04 |
| Hsp70Bbb | FBpp0082105 | FBgn0051354 | Q9VG58 | 7.0 ±2.1E−02 | 1.40E−03 | 6.9±6.0E−02 | 1.50E−03 |
| SRPK | FBpp0086515 | FBgn0026370 | Q0E965 | 7.0±6.1E−01 | 2.60E−02 | 7.6±2.8E−01 | 1.40E−02 |
| Unc-76 | FBpp0070360 | FBgn0040395 | Q9W525 | 7.0±9.5E−02 | 8.80E−03 | 6.3±8.1E−02 | 1.20E−02 |
| GstO3 | FBpp0076348 | FBgn0035904 | Q9VSL2 | 6.9±2.1E−01 | 8.40E−02 | 7.8±9.1E−02 | 4.30E−02 |
| Srp54k | FBpp0076872 | FBgn0010747 | Q9V3D9 | 6.9±5.0E−01 | 1.90E−01 | 6.4±5.3E−01 | 1.30E−01 |
| CG32069 | FBpp0075970 | FBgn0052069 | Q9VTE1 | 6.7±1.7E−01 | 1.60E−04 | 6.6±6.5E−02 | 1.70E−04 |
| alphaTub85E | FBpp0081565 | FBgn0003886 | P06604 | 6.7±7.2E−02 | 1.80E−03 | 7.1±1.0E−01 | 9.20E−04 |
| MED30 | FBpp0072457 | FBgn0035149 | Q9W0P3 | 6.6±7.5E−02 | 1.40E−04 | 6.6±6.6E−02 | 1.80E−04 |
| Fwe | FBpp0075297 | FBgn0261722 | Q95T12 | 6.6±9.6E−02 | 1.60E−04 | 6.4±8.1E−02 | 1.30E−04 |
| Rrp4 | FBpp0072020 | FBgn0034879 | Q9W1M9 | 6.4±1.1E−01 | 1.70E−03 | 6.3±6.4E−02 | 1.30E−03 |
| CG1789 | FBpp0071194 | FBgn0030063 | Q9W3C0 | 6.3±2.1E−02 | 1.10E−03 | 6.8±2.7E−02 | 7.30E−04 |
| qkr58E-2 | FBpp0071739 | FBgn0022985 | Q9W254 | 6.3±9.7E−02 | 1.80E−04 | 6.1±6.8E−02 | 2.10E−04 |
| prod | FBpp0085785 | FBgn0014269 | Q7JNE1 | 6.3±1.4E−01 | 1.20E−04 | 6.2±1.1E−01 | 1.50E−04 |
| Tsp42Ed | FBpp0085513 | FBgn0029507 | Q7JWV7 | 6.3±1.0E−01 | 2.00E−04 | 6.1±8.1E−02 | 1.90E−04 |
| CG6180 | FBpp0079979 | FBgn0032453 | Q9VK60 | 6.3±1.4E−01 | 9.20E−04 | 6.6±2.8E−02 | 6.70E−04 |
| CG7945 | FBpp0075353 | FBgn0036505 | Q95RY2 | 6.3±4.2E−01 | 1.80E−01 | 5.1±1.0E+00 | 2.80E−01 |
| CG14695 | FBpp0081811 | FBgn0037850 | Q9VGV0 | 6.2±1.9E−01 | 2.30E−03 | 6.5±1.1E−01 | 1.80E−03 |
| UQCR-C1 | FBpp0082459 | FBgn0038271 | Q9VFF0 | 6.2±6.5E−02 | 4.00E−04 | 6.0±5.0E−02 | 5.00E−04 |
| ATPsynCF6 | FBpp0083824 | FBgn0016119 | Q24407 | 6.2±2.2E−01 | 1.20E−05 | 6.2±5.0E−02 | 1.30E−05 |
| BEAF-32 | FBpp0086571 | FBgn0015602 | Q7JN06 | 6.1±5.9E−02 | 5.30E−04 | 6.2±7.5E−02 | 5.70E−04 |
| CG5021 | FBpp0290362 | FBgn0035944 | Q8IQC1 | 6.1±6.2E−01 | 1.90E−01 | 4.9±1.2E+00 | 3.10E−01 |
| PH4alphaEFB | FBpp0085012 | FBgn0039776 | Q9VA69 | 6.0±1.0E−01 | 1.70E−03 | 6.3±9.6E−02 | 1.50E−03 |
| Ns3 | FBpp0070284 | FBgn0266284 | Q9W590 | 6.0±9.2E−02 | 2.60E−01 | 6.0±7.1E−02 | 2.10E−01 |
| CG1265 | FBpp0073084 | FBgn0035517 | Q9VZF3 | 6.0±1.5E−01 | 9.70E−04 | 6.0±5.0E−02 | 1.10E−03 |
| CG12343 | FBpp0087342 | FBgn0033556 | Q9V5Q4 | 6.0±6.5E−02 | 1.20E−04 | 6.0±6.7E−02 | 1.10E−04 |
| CG11030 | FBpp0078766 | FBgn0031736 | Q9VMM7 | 5.9±3.8E−01 | 3.00E−01 | 5.6±4.3E−01 | 2.40E−01 |
| Hmg-2 | FBpp0071501 | FBgn0026582 | Q9W2K8 | 5.9±8.6E−02 | 1.80E−04 | 5.7±9.0E−02 | 2.50E−04 |
| Prx5 | FBpp0100079 | FBgn0038570 | Q960M4 | 5.9±1.2E−01 | 2.00E−02 | 6.1±4.6E−02 | 1.60E−02 |
| Jheh2 | FBpp0088908 | FBgn0034405 | Q7KB18 | 5.9±6.5E−02 | 8.40E−04 | 5.9±1.1E−02 | 8.20E−04 |
| Pcd | FBpp0084824 | FBgn0024841 | O76454 | 5.9±6.6E−02 | 4.70E−03 | 6.3±1.1E−01 | 3.50E−03 |
| caz | FBpp0073996 | FBgn0011571 | Q27294 | 5.7±1.3E−01 | 2.10E−05 | 6.0±6.1E−02 | 8.70E−06 |
| Sgf29 | FBpp0071543 | FBgn0050390 | Q9W2I4 | 5.7±1.9E−01 | 1.80E−04 | 5.6±8.5E−02 | 2.40E−04 |
| vnc | FBpp0076077 | FBgn0263251 | Q9VT75 | 5.6±4.8E−02 | 1.80E−03 | 6.0±1.4E−01 | 1.40E−03 |
| Dbp45A | FBpp0087645 | FBgn0010220 | Q07886 | 5.6±2.3E−01 | 1.30E−03 | 6.0±3.1E−02 | 1.00E−03 |
| Uch-L5 | FBpp0076200 | FBgn0011327 | Q9XZ61 | 5.5±1.4E−01 | 7.70E−04 | 5.7±6.9E−02 | 6.00E−04 |
| Fmr1 | FBpp0081675 | FBgn0028734 | Q9NFU0 | 5.5E±4.5E−02 | 2.00E−01 | 5.4±1.2E−01 | 1.70E−01 |
| gfzf | FBpp0290855 | FBgn0250732 | Q6NP69 | 5.5±9.0E−02 | 2.70E−04 | 5.5±1.1E−01 | 2.50E−04 |
| su(Hw) | FBpp0082404 | FBgn0003567 | P08970 | 5.4±8.7E−02 | 6.00E−04 | 5.3±8.2E−02 | 4.10E−04 |
| CG6084 | FBpp0075870 | FBgn0086254 | Q9VTK9 | 5.4±9.2E−02 | 7.80E−03 | 5.6±4.8E−02 | 6.70E−03 |
| Nup50 | FBpp0087861 | FBgn0033264 | Q7K0D8 | 5.4±2.8E−02 | 1.10E−01 | 4.1±3.3E−01 | 2.10E−01 |
| RagC-D | FBpp0087883 | FBgn0033272 | Q7K519 | 5.4±1.5E−01 | 5.40E−04 | 5.3±1.6E−01 | 4.00E−04 |
| CG17271 | FBpp0083397 | FBgn0038829 | Q9VDI5 | 5.4±6.9E−01 | 2.50E−01 | 4.3±1.2E+00 | 1.80E−01 |
| Caf1 | FBpp0082511 | FBgn0263979 | Q24572 | 5.3±1.7E−07 | 7.20E−03 | 5.3±6.8E−07 | 6.50E−03 |
| CG1091 | FBpp0081154 | FBgn0037470 | Q9VI58 | 5.3±1.2E−01 | 7.20E−04 | 5.3±1.3E−01 | 9.30E−04 |
| bor | FBpp0082728 | FBgn0040237 | Q9VEX6 | 5.1±1.2E−01 | 2.80E−02 | 4.2±4.1E−01 | 6.80E−02 |
| Spt6 | FBpp0070861 | FBgn0028982 | Q9W420 | 5.1±3.9E−02 | 1.90E−01 | 4.8±7.6E−02 | 1.90E−01 |
| YT521-B | FBpp0072940 | FBgn0027616 | Q9VZQ1 | 5.0±3.5E−01 | 3.50E−01 | 5.0±9.7E−02 | 3.20E−01 |
| CG5482 | FBpp0085842 | FBgn0034368 | Q7K3D4 | 5.0±9.0E−02 | 3.40E−04 | 5.3±5.3E−02 | 2.20E−04 |
| CG1309 | FBpp0073085 | FBgn0035519 | Q9VZF1 | 4.9±7.4E−03 | 8.90E−03 | 5.1±3.7E−02 | 4.00E−03 |
| hyd | FBpp0081568 | FBgn0002431 | P51592 | 4.9±1.6E−01 | 1.80E−03 | 4.8±4.4E−02 | 2.30E−03 |
| CG32344 | FBpp0072475 | FBgn0052344 | Q8SY39 | 4.9±5.8E−06 | 4.20E−01 | 11.1±4.1E−02 | 3.10E−02 |
| PQBP1 | FBpp0084140 | FBgn0039270 | Q9VBY6 | 4.9±2.9E−01 | 6.90E−02 | 5.8±2.5E−01 | 3.30E−02 |
| ATPsynO | FBpp0082522 | FBgn0016691 | Q24439 | 4.8±1.0E−01 | 2.30E−02 | 5.1±7.6E−02 | 1.90E−02 |
| CG4858 | FBpp0077868 | FBgn0037011 | Q9VPD2 | 4.8±1.5E−01 | 6.70E−01 | 6.5±5.8E−02 | 3.30E−01 |
| CG3335 | FBpp0076112 | FBgn0036018 | Q9VT19 | 4.8±9.8E−02 | 5.80E−03 | 5.0±7.8E−02 | 4.20E−03 |
| CG11076 | FBpp0088285 | FBgn0039929 | Q9V493 | 4.8±4.3E−02 | 2.30E−03 | 4.7±1.0E−01 | 2.10E−03 |
| YL-1 | FBpp0079735 | FBgn0032321 | Q9VKM6 | 4.7±8.9E−02 | 6.90E−04 | 4.7±5.9E−02 | 7.10E−04 |
| sub | FBpp0086041 | FBgn0003545 | Q9V877 | 4.6±1.0E−01 | 5.30E−03 | 4.6±1.3E−01 | 6.70E−03 |
| Rbcn-3A | FBpp0292404 | FBgn0023458 | Q9W425 | 4.6±1.4E−01 | 3.60E−04 | 4.5±7.8E−02 | 4.10E−04 |
| eIF-4B | FBpp0112403 | FBgn0020660 | Q7PLL3 | 4.6±8.0E−02 | 1.90E−02 | 4.8±8.2E−02 | 1.30E−02 |
| Pgi | FBpp0087760 | FBgn0003074 | P52029 | 4.5±9.7E−02 | 9.70E−03 | 4.7±1.6E−02 | 7.20E−03 |
| CG9630 | FBpp0081355 | FBgn0037561 | Q9VHU1 | 4.5±7.3E−02 | 2.80E−03 | 4.8±1.6E−01 | 1.20E−03 |
| Cenp-C | FBpp0088911 | FBgn0266916 | Q9VHP9 | 4.5±1.3E−01 | 3.00E−03 | 4.4±7.0E−02 | 2.90E−03 |
| Mtr3 | FBpp0074687 | FBgn0036916 | Q9VW53 | 4.5±1.1E−01 | 5.20E−01 | 5.8±6.4E−02 | 2.70E−01 |
| CG2943 | FBpp0081262 | FBgn0037530 | Q9VHY6 | 4.5±7.7E−02 | 6.10E−01 | 4.4±5.5E−02 | 5.10E−01 |
| Cdk12 | FBpp0078013 | FBgn0037093 | Q9VP22 | 4.4±2.2E−01 | 5.00E−04 | 4.2±7.5E−02 | 5.70E−04 |
| CG7518 | FBpp0297366 | FBgn0038108 | A0A0B4K6G6 | 4.3±1.2E−01 | 5.40E−04 | 4.4±7.4E−02 | 6.00E−04 |
| holn1 | FBpp0079616 | FBgn0032250 | Q9VKV5 | 4.2±3.8E−02 | 3.20E−02 | 4.6±1.2E−01 | 1.10E−02 |
Fig. 2.
Predicted associations between centromere associated proteins. Confidence view of protein associations among the 86 centromere enriched proteins predicted with the STRING web-tool. Stronger associations are represented by thicker lines.
2. Materials and methods
2.1. Cell culture
The Drosophila Schneider Line 2 derived L2–4 cell line was used for all experiments. Cells were maintained at 25 °C in Drosophila Schneider medium supplemented with 10% fetal calf serum and penicillin/streptomycin. Stable cell lines were established by XtremeGENE HP mediated transfection of GFP-fusion expression constructs following four weeks of Hygromycin B selection (100 µg/mL).
2.2. Chromatin preparation for AP-MS
Asynchronously growing cells were harvested by centrifugation and washed in PBS. Cells were resuspended in ice-cold hypotonic buffer (10 mM HEPES, pH 7.6; 15 mM NaCl; 1.5 mM MgCl2; 0.1 mM DTT; freshly added protease inhibitors: PMSF, Aprotinin, Leupeptin, Pepstatin) and lysed for 10 min on ice by adding Triton X-100 to a final concentration of 0.1%. Nuclei were pelleted by centrifugation, washed with PBS and chromatin was solubilized for 10 min at 26 °C by micrococcal nuclease (MNase) digestion in EX100 buffer (10 mM HEPES, pH 7.6; 100 mM NaCl; 1.5 mM MgCl2; 0.5 mM EGTA; 2 mM CaCl2; 10% glycerol (v/v); freshly added protease inhibitors) containing 2000 U MNase per one billion nuclei. Chromatin was released by increasing the sodium chloride concentration to 300 mM and applying ten strokes in a Dounce homogenizer with a tight-fit pestle. Following one hour incubation at 4 °C, insoluble material was pelleted for 20 min at 20,000g and the supernatant was precleared with Protein A Sepharose beads yielding the AP-MS input extract.
2.3. Affinity purification and sample preparation for mass spectrometry
GFP-Trap agarose beads (ChromoTek) were used as affinity resin. The beads were preblocked in 0.5% BSA, 0.5% polyvinylpyrrolidone dissolved in EX100 buffer by over-head rotation for 30 min. The input extract was added to the blocked beads and affinity purification was performed for 2 h at 4 °C on an over-head rotator. The affinity resin with bound complexes was washed three times for 5 min at 4 °C with EX300 buffer and bound proteins were eluted by boiling beads in Laemmli buffer for 10 min at 95 °C. Eluted proteins were separated by SDS-PAGE using a 15% polyacrylamide gel and the gel was stained with Coomassie Brilliant Blue G-250. Whole lanes were excised from the gel with a disposable gridcutter (Gel Company) and split into eight vials. Following destaining, reduction of disulfide bonds with dithiothreitol and alkylation with iodoacetamide, in-gel tryptic digestion was performed. Resulting peptides were collected by acid extraction of the gel pieces, concentrated by evaporation, and resuspended in 0.1% TFA.
2.4. LC–MS/MS
Peptides were injected into an Ultimate 3000 HPLC system (Thermo-Fisher Scientific). Samples were desalted online by a C18 micro-column (5 mm×300 µm id 5 mm, packed with C18 PepMapTM, 5 µm, 100 Å, Thermo-Fisher Scientific), and peptides were separated with a gradient from 5% to 60% acetonitrile in 0.1% formic acid over 40 min at 300 nL/min on a C18 analytical column (10 cm×75 µm, packed in house with C18 PepMapTM, 3 µm, 100 Å, Thermo-Fisher Scientific). The effluent from the HPLC was directly infused into the LTQ Orbitrap mass spectrometer (Thermo-Fisher Scientific) via a nano-electrospray ion source. The MS instrument was operated in the data-dependent mode to automatically switch between full-scan MS and MS/MS acquisition. Survey fullscan MS spectra (m/z 350–2000) were acquired in the Orbitrap with resolution 60,000 at m/z 400. For all measurements with the Orbitrap detector, three lock-mass ions from ambient air (m/z=371.10123, 445.12002, 519.13882) were used for internal calibration as described [4]. The six most intense peptide signals with charge states between two and five were sequentially isolated applying a 1 Da window centered around the most abundant isotope to a target value of 10,000 and fragmented in the linear ion trap by collision-induced dissociation. Fragment ion spectra were recorded in the linear trap of the instrument. Typical mass spectrometric conditions were as follows: spray voltage, 1.4 kV; no sheath and auxiliary gas flow; heated capillary temperature, 200 °C; activation time, 30 ms; and normalized collision energy, 35% for collision-induced dissociation in linear ion trap.
2.5. Protein identification and statistical analysis
For protein identification, the raw data were analyzed with the Andromeda algorithm of the MaxQuant protein analysis package (version 1.2.2.5) against the Flybase dmel-all-translation-r5.24.fasta database including reverse sequences and contaminants. The Trypsin/P enzyme was selected, allowing for maximum two missed cleavages. Carbamidomethylation of cysteine was set as fixed modification; methionine oxidation and protein N-acetylation were included as variable modifications. The mass tolerance of the initial search was 20 ppm; after recalibration, a 6 ppm mass error was applied for the main search. Fragment ions were searched with a mass offset of 0.5 Da using the six most intense signals per 100 Da. Searching for secondary peptide hits within already assigned MS/MS spectra was enabled. The search results were filtered with a peptide and protein false discovery rate of 0.01 with a minimum peptide length of six amino acids. Protein identifications with at least one unique peptide were accepted. For quantification, the intensity-based absolute quantification (iBAQ) values were calculated from peptide intensities and the protein sequence information [5] of unmodified, M/oxidated, and acetylated peptide species with a minimum of two peptides per protein.
As preparation for statistical analysis, protein hits representing reversed sequences or contaminants and protein hits without quantification values were removed from the list of identified proteins from three biological replicates. iBAQ quantification values were log2-transformed and subsequently missing values were imputed from a random distribution centered at 1/3×log2 of the obtained experimental data. The imputation was repeated three times to reduce effects of the random value distribution. ANOVA was applied in DanteR (vs 0.2, PNNL, Richland, WA, USA) [6] to calculate protein enrichment factors and p-values and obtained p-values were corrected for multiple hypothesis testing by the Benjamini–Hochberg method [7].
2.6. STRING analysis
Protein names from Table 1 were subjected to STRING analysis using the web-tool available via http://string-db.org/ [3]. Fig. 2 shows the confidence view with the active prediction methods “Experiments”, “Databases” and “Textmining” and medium confidence score (0.4).
Acknowledgments
We would like to thank all members of the Imhof and Heun group for critical discussions and feedback and the anonymous reviewer for valuable comments and suggestions to improve the manuscript. Work in the lab of A.I. was supported by grants from the German Research Council (Deutsche Forschungsgemeinschaft (DFG; Projekt IM 23/9-1)) and the European Union (EpiGeneSys, 257082). Work in the lab of P.H. was sponsored by an ERC (ERC-2012-StG_20111109) grant (ERC-BioSynCen).
References
- 1.T.K. Barth, G.O. Schade, A. Schmidt, I. Vetter, M. Wirth, P. Heun, et al. Identification of novel Drosophila centromere-associated proteins. Proteomics 14 (2014) 2167-2178. [DOI] [PubMed]
- 2.Erhardt S., Mellone B.G., Betts C.M., Zhang W., Karpen G.H., Straight A.F. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen L.J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J. STRING 8 – a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsen J.V., de Godoy L.M., Li G., Macek B., Mortensen P., Pesch R. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteom. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.B. Schwanhausser, D. Busse, N. Li, G. Dittmar, J. Schuchhardt, J. Wolf, et al. Global quantification of mammalian gene expression control. Nature.473:337-342. [DOI] [PubMed]
- 6.T. Taverner, Y.V. Karpievitch, A.D. Polpitiya, J.N. Brown, A.R. Dabney, G.A. Anderson, et al. DanteR: an extensible R-based tool for quantitative analysis of -omics data. Bioinformatics.28:2404-2406. [DOI] [PMC free article] [PubMed]
- 7.Benjamini Y., Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B: Methodol. 1995;57:289–300. [Google Scholar]