Rodriguez-Barrueco et al. found that HR−/HER2+ cells secrete high levels of IL-6, inducing the activation of STAT3, which in turn promotes a second autocrine stimulus to increase S100A8/9 complex (calprotectin) production and secretion. Inhibition of the IL-6–JAK2–STAT3–calprotectin axis with FDA-approved drugs, alone and in combination with HER2 inhibitors, reduced the tumorigenicity of HR−/HER2+ breast cancers.
Keywords: genetic screen, HER2, STAT3, tailored therapies, breast cancer
Abstract
HER2-positive (HER2+) breast adenocarcinomas are a heterogeneous group in which hormone receptor (HR) status influences therapeutic decisions and patient outcome. By combining genome-wide RNAi screens with regulatory network analysis, we identified STAT3 as a critically activated master regulator of HR−/HER2+ tumors, eliciting tumor dependency in these cells. Mechanistically, HR−/HER2+ cells secrete high levels of the interleukin-6 (IL-6) cytokine, inducing the activation of STAT3, which in turn promotes a second autocrine stimulus to increase S100A8/9 complex (calprotectin) production and secretion. Increased calprotectin levels activate signaling pathways involved in proliferation and resistance. Importantly, we demonstrated that inhibition of the IL-6–Janus kinase 2 (JAK2)–STAT3–calprotectin axis with FDA-approved drugs, alone and in combination with HER2 inhibitors, reduced the tumorigenicity of HR−/HER2+ breast cancers, opening novel targeted therapeutic opportunities.
HER2/ERBB2 is a receptor tyrosine kinase found overexpressed in 15%–20% of breast tumors (HER2+ tumors) (Hynes and Lane 2005). It belongs to a family of four receptors (EGFR/HER1, HER2, HER3, and HER4) involved in cell signaling networks regulating cell growth, survival, and differentiation (Hynes and Lane 2005; Lemmon and Schlessinger 2010). In general, EGFR/HER receptors are activated through dimerization, which is promoted by the binding of cognate growth factors. However, HER2 is a unique member of the family and does not rely on direct ligand binding for activation; instead, it is activated through heterodimerization with other EGFR/HER family members that are ligand-bound (Citri and Yarden 2006). When aberrantly overexpressed, HER2 is able to homodimerize and initiate proliferation and prosurvival signal transduction in a ligand-independent fashion (Citri and Yarden 2006).
Despite the existence of several HER2-specific targeted therapies that have drastically improved the treatment of HER2+ patients (trastuzumab, lapatinib, TDM1, and pertuzumab), tumor progression remains transient (Hynes and Lane 2005; Tagliabue et al. 2010). Thus, there is a need to find complementary, therapeutic targets that hold the potential for more effective treatments.
Although, HER2+ tumors are commonly considered as a single entity, there is increasing evidence indicating that important intrinsic differences associated with hormone receptor (HR) status exist. Each of the two groups, HR+ and HR−, represents about half of all HER2+ breast cancers.
Compared with HR+/HER2+, HR−/HER2+ tumors present worse histopathological characteristics (larger size, lymph node involvement, higher American Joint Committee on Cancer [AJCC] stage, and higher histological grade) (Vaz-Luis et al. 2012). HR+/HER2+ tumors preferentially recur in bones, while there is a strong trend for more visceral metastases in the HR−/HER2+ cancers (Paluch-Shimon et al. 2009; Vaz-Luis et al. 2012). Moreover, despite a higher rate of pathologic complete responses (pCRs) to neoadjuvant chemotherapy (Hurley et al. 2006; Vaz-Luis et al. 2012), HR−/HER2+ patients still have an increased risk of death within 5 years of diagnosis (Vaz-Luis et al. 2012). Intrinsic differences between HR−/HER2+ and HR+/HER2+ breast cancers are also found at the molecular level, as highlighted by unsupervised cluster analysis of gene expression profiles. The latter clearly identifies two distinct HER2+ subtypes. Most tumors clinically classified as HR+/HER2+ fall in the luminal B subtype, while most HR−/HER2+ tumors are part of the HER2-enriched subtype (Perou et al. 2000; Sotiriou and Pusztai 2009).
Indeed, while HR+/HER2+ patients benefit from anti-hormonal and HER2 targeted therapies (Kaufman et al. 2009), the outcome for HR−/HER2+ patients strongly depends on their response to chemotherapy as well as anti-HER2 therapy. Thus, to identify genes that represent novel mechanistic dependencies in HR−/HER2+ breast cancer cells, we designed an integrative approach that combines functional genomic (RNAi screens) (Luo et al. 2008; Silva et al. 2008) and computational (Basso et al. 2005; Margolin et al. 2006; Carro et al. 2010; Lefebvre et al. 2010; Piovan et al. 2013) algorithms.
Our integrative analysis identified STAT3 as a de novo master regulator (MR) gene associated with HER2-mediated transformation in HR− breast cancer cells. Importantly, we demonstrate that aberrant STAT3 activity is necessary to maintain the HR−/HER2+ tumor state, thus representing a nononcogene dependency in these tumors. Mechanistically, we found that HR−/HER2+ breast tumors secrete high levels of interleukin-6 (IL-6). This autocrine mechanism induces the activation of STAT3 via the canonical Janus kinase 2 (JAK2)/STAT3 pathway. Aberrant STAT3 activity induces up-regulation and secretion of the S100A8/9 complex (calprotectin), thus triggering a second autocrine stimulus that enhances proliferation and survival. As a result, disruption of the “IL-6–JAK2–STAT3–S100A8/9 cascade” compromises HR−/HER2+ cell viability, suggesting that the components of this pathway represent putative therapeutic targets in HR−/HER2 tumors. Importantly, small molecule inhibitors and blocking antibodies for components of this double autocrine loop are already FDA-approved or in clinical trials. Here, we demonstrate that blocking the IL-6 receptor (IL-6R) with the humanized monoclonal antibody tocilizumab (Patel and Moreland 2010; Navarro-Millan et al. 2012), STAT3 inactivation with the dual JAK1/2 inhibitor ruxolitinib (Mascarenhas and Hoffman 2012; Mesa et al. 2012), or calprotectin inhibition with the small molecule inhibitor tasquinimod (Dalrymple et al. 2007, 2012), alone or in combination with anti-HER2 therapies, compromises the viability of HR−/HER2+ breast cancer cells. The availability of FDA-approved inhibitors to target this novel mechanism represents an exciting opportunity for rapid translation of these findings to the clinics.
Results
Genome-wide RNAi screens and MR analysis identify STAT3 as an essential MR gene in HR−/HER2+ cells
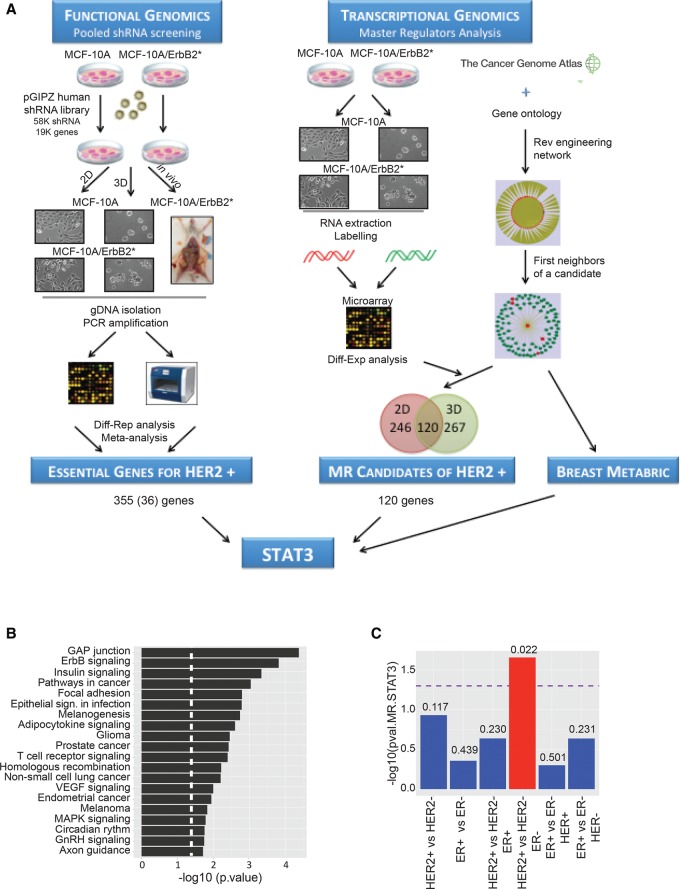
To identify critical regulators of HR−/HER2+ breast cancer cell homeostasis, we developed an integrative approach that combines pooled, genome-wide loss-of-function studies and systems biology methods for the reverse engineering and interrogation of regulatory networks (Fig. 1A).
Figure 1.
Integrative functional studies identify STAT3 as a MR in HR−/HER2+ breast cancer cells. (A) Schematic representation of the strategy followed to identify potential therapeutic targets in HR−/HER2+ breast tumors as described in the text. The left side shows a simplified representation of the shRNA screening strategy. The right side summarizes MR analysis based on expression profiles from MCF-10A and MCF-10A/ErbB2* models as well as from primary samples (TCGA [The Cancer Genome Atlas Network] and METABRIC [Molecular Taxonomy of Breast Cancer International Consortium]) (for a detailed description, see the Materials and Methods). The integration of both approaches identified STAT3 as a putative target for HR−/HER2+ breast cancer cells. (B) Enrichment pathway analysis of the genes that have a stronger inhibitory effect on the growth of MCF-10A/ErbB2* cells when compared with MCF-10A. (C) Correlation of STAT3 activity (MR score) with the breast cancer subtype defined by HER2 and HR status in primary samples from the METABRIC data set. The number shown on the bars corresponds to the P-value for each correlation, and the dashed line indicates the significance threshold.
RNAi is a powerful genetic strategy to interrogate gene function by loss-of-function studies. We (Silva et al. 2008; Rodriguez-Barrueco et al. 2013; Yu et al. 2013) and others (Luo et al. 2008; Schlabach et al. 2008) have developed RNAi-based genetic approaches to perform high-throughput (HTP) screens in mammalian systems. Using this technology, we performed genome-wide pooled RNAi screens to find gene functions that are required to maintain the homeostasis of HR− mammary epithelial cells transformed by the oncogene HER2. However, RNAi-based screens suffer from high inherent noise and heterogeneity that can compromise the effective validation of identified hits (Schultz et al. 2011). To reduce the candidate gene list size while increasing its robustness, we decided to combine our genetic screens with computational approaches that are highly effective in identifying MR genes of HR−/HER2+ tumors. These represent transcription factors (TFs) and signaling proteins (SPs) that are mechanistic regulators of the genes that are differentially expressed between normal and transformed cells from a repertoire of 780 expressed TFs and 2453 expressed SPs that are expressed in HR−/HER2+ tumors. This is accomplished by using MARINa (Master Regulator Inference Algorithm) (Carro et al. 2010; Lefebvre et al. 2010; Piovan et al. 2013) to interrogate regulatory networks (hereafter referred to as interactomes) inferred by ARACNe (Algorithm for the Reconstruction of Accurate Cellular Networks) (Basso et al. 2005; Margolin et al. 2006) from a large, tumor-specific gene expression profile data set. Importantly, we showed that 20%–30% of the MARINa-inferred MRs are essential for viability of the cells that they regulate (Carro et al. 2010; Piovan et al. 2013), thus representing valuable tumor-specific targets. We speculated that inhibiting MRs that are computationally identified and also selected by the loss-of-function screen would impact HER2-mediated transformation of HR− mammary epithelial cells.
As the experimental model for the RNAi screens, we chose mammary epithelial MCF-10A cells and an isogenic variant transformed with an oncogenic form of HER2 (Wang et al. 2006), termed here MCF-10A/ErbB2*. This model system possesses several characteristics that make it useful for our purposes. MCF-10A is a spontaneously immortalized, but nontransformed, human mammary epithelial cell line (Soule et al. 1990; Debnath et al. 2003). These cells exhibit numerous features of normal breast epithelium, including diploid chromosome genotype, lack of tumorigenicity in nude mice, lack of anchorage-independent growth, and dependence on growth factors for proliferation and survival. In contrast, MCF-10A/ErbB2* cells are transformed and form tumors when xenografted into immunocompromised mice. Importantly, parental MCF-10A cells are HR−, and, when grown on basement membrane components (BCMs), these cells activate a morphogenetic program that induces the development of organotypic spheroid structures. Remarkably, this three-dimensional (3D) model bears a much closer resemblance to the in vivo epithelial context than classical two-dimensional (2D) cultures (Debnath et al. 2002, 2003).
Using this cell model, we completed three complementary RNAi screens (Fig. 1A). In the first screen, wild-type MCF-10A and MCF-10A/ErbB2* cells were grown on standard tissue culture plates. In the second screen, these cells were grown in the presence of BCM (extracellular matrix [ECM]-Matrigel) to form organotypic structures. Finally, we performed an in vivo screen by transplanting MCF-10A/ErbB2* cells into the mammary fat pads of immunocompromised (SCID) mice. We reasoned that knockdown of genes that specifically compromised the viability of HER2 transformed cells in all screens may represent strong tumor dependencies that are conserved independently of the conditions and platform. Metaanalysis identified 355 genes fulfilling this criterion. Thirty-six of them were TFs and/or signaling molecules (Supplemental Table S1). Pathway analysis showed that, as expected, silencing components of HER2 as well as canonical MAPK signaling have a stronger impact on the growth of MCF-10A/ErbB2* cells when compared with nontransformed MCF-10A (Fig. 1B; Supplemental Fig. S1A,B).
We then used MARINa to computationally identify candidate MRs of HER2-mediated transformation of MCF-10A/ErbB2* breast cancer cells (Fig. 1A). First, we analyzed 359 gene expression profiles from TCGA (The Cancer Genome Atlas Network) primary breast cancer samples (The Cancer Genome Atlas Network 2012) to construct breast cancer-specific regulatory (11,415 nodes and 112,490 edges) and signaling (11,477 nodes and 180,220 edges) networks using ARACNe, an information theory-based reverse-engineering approach (Basso et al. 2005; Margolin et al. 2006). For SPs, we used ARACNe to identify their maximum information transfer (MIT) targets, as previously done for the identification of AKT1 as the MR of glucocorticoid resistance in T acute lymphoblastic leukemia (T-ALL) (Piovan et al. 2013). We then interrogated these interactomes with genes differentially expressed in the MCF-10A versus MCF-10A/ErbB2* cells (in both 2D and 3D cultures) (see Supplemental Table S2) using MARINa to identify the transcriptional (Carro et al. 2010; Lefebvre et al. 2010; Aytes et al. 2014) and signaling (Piovan et al. 2013) MRs that are mechanistic regulators of HER2-dependent tumorigenesis in HR− cells. These analyses showed significant overlap of MARINa-inferred MRs between the 2D and 3D experimental models (Fig. 1A). In fact, there was a dramatic increase compared with the poor overlap of signature genes, by conventional differential expression analysis (Supplemental Fig. S1C). This suggests that, while the programs that affect HER2-mediated transformation in these two contexts are substantially different, their transcriptional and signaling regulators are highly conserved.
Integration of 120 MRs predicted by systems biology analysis and 36 depleted candidates from metaanalysis of RNAi screens identified STAT3, AGRN, and GLRX as potential MRs of MCF-10A/ErbB2* tumorigenesis and thus as candidate therapeutic targets (Supplemental Fig. S1D). We further validated STAT3 and AGRN as essential genes in MCF-10A/ErbB2* cells using additional shRNAs that efficiently silence these genes but were not included in the initial pool library (Supplemental Fig. S1E). Individual gene validation was performed through competition assays. These assays are based on the ability of the shRNA vectors to coexpress the shRNA of interest and a fluorescent protein. Cells expressing shRNAs were mixed in a 50/50 ratio with parental cells, which did not express the hairpin; expression of the fluorescent reporter allowed us to track the ratio of shRNA/non-shRNA cells by flow cytometry (Supplemental Fig. S1E). Interestingly, STAT3 and AGRN showed no differential expression in either 2D or 3D, and conventional differential expression analysis could not identify them; however, MARINa analysis successfully inferred them as differentially activated (Supplemental Fig. S1D). This is a recurrent theme, as virtually all validated MRs in other tumor contexts were not differentially expressed (Carro et al. 2010; Piovan et al. 2013).
Next, we evaluated the MR activity of STAT3 and AGRN in primary breast cancer samples. To avoid overfitting, we used a METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) data set (Curtis et al. 2012) independent of the TCGA data from which the networks were derived. When we compared all major clinical subtypes based on HR and HER2 status, STAT3 was the only one of the three MRs that consistently emerged as a HR−/HER2+-specific MR (Fig. 1C; Supplemental Fig. S1F).
STAT3 silencing compromised HR−/HER2+ breast cancer cell viability in vitro and in vivo
An increasing amount of evidence suggests a pivotal role for STAT3 as a regulator of cancer cell homeostasis (Darnell 2005; Weerasinghe et al. 2007). This includes breast cancers, where high levels of phosphorylated STAT3 (p-STAT3) are found in ∼50% of cases and are associated with high expression levels of growth factor receptors EGFR and HER2 (Berclaz et al. 2001; Berishaj et al. 2007; Hartman et al. 2011). The canonical JAK/STAT pathway consists of a series of membrane receptors that bind to a family of cytoplasmic kinases, the JAKs, for signal transduction (Rawlings et al. 2004). Association of the receptor with its respective cytokine/ligand enables the transduction of the intracellular signal by phosphorylating and activating TFs called STATs.
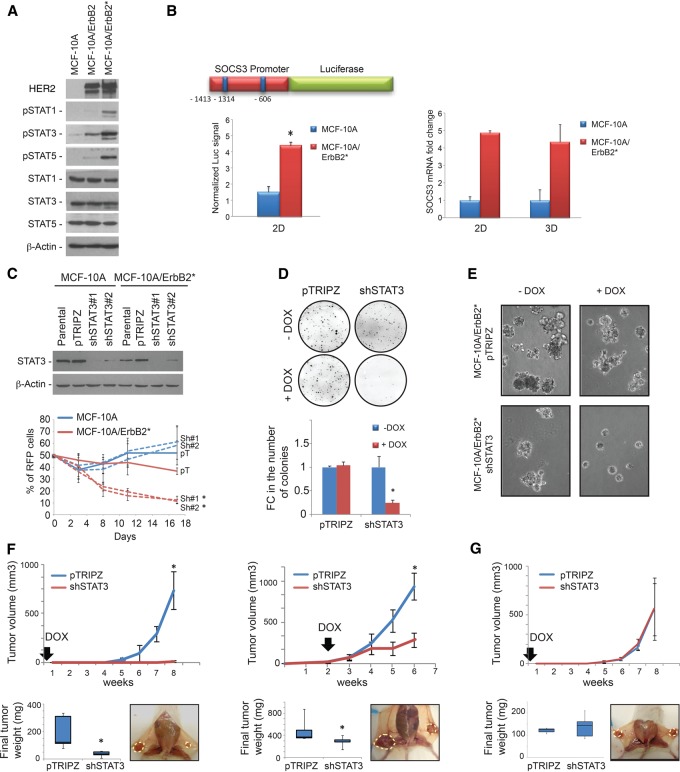
As shown by interactome analysis and RNAi screens, multiple evidences suggest that the JAK/STAT pathway is activated in HER2 transformed MCF-10A cells. Indeed, we found that STAT1, STAT3, and STAT5 phosphorylation increased upon overexpression of both wild-type ErbB2 and the mutant ErbB2* variant used in our screens (Fig. 2A; Wang et al. 2006). Additionally, a reporter assay in which luciferase expression was controlled by the promoter of the bona fide endogenous STAT3 target SOCS3 (Wormald and Hilton 2004) showed a more than fourfold increase in MCF-10A/ErbB2* compared with parental MCF-10A (Fig. 2B). Consistently, we found that endogenous SOCS3 mRNA levels increased (fivefold) in ErbB2* transformed MCF-10A cells in both 2D and 3D cultures (Fig. 2B). Critically, STAT3 activity was exclusively linked to increased phosphorylation and not increased expression (Fig. 2A). Remarkably, robust activation of STAT3 was observed when MCF-10A cells were transformed with HER2 but not any common breast cancer drivers (Supplemental Fig. S2A). Overall, these data demonstrate that HER2 overexpression selectively activates the JAK/STAT pathway.
Figure 2.
RNAi-mediated silencing of STAT3 expression reduces the tumorigenicity of HR−/HER2+ breast cancer cells. (A) Western blot analysis of STAT1, STAT3, and STAT5 phosphorylation in MCF-10A cells and its isogenic variants overexpressing ErbB2 wild type and the oncogenic ErbB2YVMA mutant (ErbB2*). (B) The left panel shows a scheme of the SOCS3 promoter subcloned upstream of the firefly luciferase reporter gene. Blue squares indicate the predicted STAT-binding sequences present in the promoter. The graph below shows the normalized luciferase signal when the SOCS3 promoter construct was transduced into MCF-10A and MCF-10A/ErbB2* cells. The right panel shows the comparison of endogenous SOCS3 expression levels between MCF-10A/ErbB2* and parental MCF-10A cells grown in plastic culture plates (2D) and Matrigel (3D). (C) Competition assay to determine the viability of MCF-10A models when STAT3 was silenced. Western blot analysis of STAT3 expression after the induction of silencing shRNAs with doxycycline (Dox) is shown in the top panel. pTRIPZ represents the control vector. The bottom panel shows the relative abundance of MCF-10A and MCF-10A/ErbB2* cells expressing control or STAT3 shRNAs (RFP+) through time after they were mixed at a 50:50 ratio with nontransduced cells at the initial time point. Dashed lines correspond to the cells where STAT3 expression has been reduced by the induction of shRNA. Colony formation assay in agar (D) and Matrigel (E) of MCF-10A/ErbB2* cells expressing Dox-inducible STAT3 shRNAs or the control vector pTRIPZ. Pictures were taken 6 d after plating of the cells. (F) Tumor growth of MCF-10A/ErbB2*-expressing control (pTRIPZ) or shRNAs against STAT3 (shSTAT3) when orthotopically injected into the intramammary fat pad (IMFP) of immunosuppressed SCID mice (N = 5). The box plot represents the weight of the tumors after their extraction. The left panel shows the results when Dox is administered in the drinking water immediately after the transplantation. The right panel shows the tumor growth when Dox is added after the tumor reached 0.3 cc (indicated by the arrows). (G) Tumor growth and final weight when MDA-MB-231 cells are othotopically injected into the IMFP of SCID mice and Dox is administered from the day of surgery. n = 5. (*) P < 0.05.
Next, we further validated STAT3 essentiality in MCF-10A/ErbB2* cells. To prevent uncontrolled cell viability loss when silencing STAT3, we opted for a doxycycline (Dox)-inducible RNAi system. This system coexpresses the shRNA of interest (shRNA sequence from the pool screens) and a red fluorescent protein (RFP) under controlled conditions (addition of Dox to the culture medium). The expression of the fluorescent reporter was used for tracking shRNA-expressing cells in competition assays, as described above (Supplemental Fig. S1E). As before, competition assays showed a progressive loss of MCF-10A/ErbB2* cells following STAT3 silencing, while no significant effect was observed in MCF-10A cells (Fig. 2C). Loss of cell viability was not limited to standard culture conditions and was also evident in in semisolid agar medium and 3D cultures (Fig. 2D,E). STAT3 silencing affected MCF-10A/ErbB2* cell growth not only in vitro but also in vivo, and STAT3 knockdown cells showed reduced tumorigenicity in orthotopic NOD scid mouse xenografts (Fig. 2F). Since parental MCF-10A cells are not tumorigenic, we used MDA-MB-231 cells to study the effect of STAT3 silencing on HER2WT cells that lack significant STAT3 activation in vivo (Supplemental Fig. S2B). As expected, tumors generated by transplanting MDA-MB-231 breast cancer cells were not affected by STAT3 knockdown (Fig. 2G; Supplemental Fig. S2C,D).
STAT3 activation in HER2+ breast cancer is mediated by autocrine secretion of IL-6 and JAK2 activation
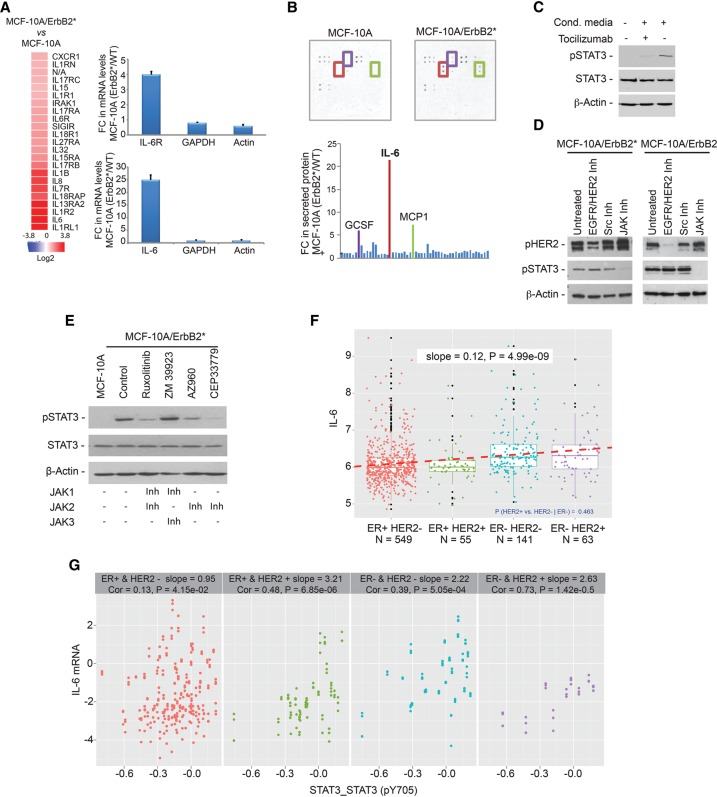
We next investigated the mechanisms that may mediate aberrant STAT3 activation. By comparing gene expression profiles of parental and HER2 transformed MCF-10A cells, we found strong up-regulation of multiple interleukin molecules and their receptors (Fig. 3A). Remarkably, the most up-regulated interleukin (>20-fold) was IL-6, a bona fide STAT3 activator (Rawlings et al. 2004; Murray 2007). High levels of IL-6 mRNA were reflected in a 21-fold increase in the levels of secreted IL-6 into the culture medium (Fig. 3B). Remarkably, incubation of MCF-10A cells with MCF-10A/ErbB2* conditioned medium strongly induced STAT3 phosphorylation, an effect that was abrogated by the presence of IL-6R-blocking antibodies (tocilizumab) (Fig. 3C; Supplemental Fig. S3A; Navarro-Millan et al. 2012). These data confirm previous observations linking HER2-mediated transformation with a profound transcriptional activation of inflammatory programs (Hartman et al. 2011).
Figure 3.
The autocrine loop IL6–JAK2 activates STAT3 in HR−/HER2+ breast cancer cells. (A, left) The heat map shows the most up-regulated ILs and IL receptors, comparing MCF-10A/ErbB2* cells and parental MCF-10A cells. (Right panel) Further validation of IL-6 and IL-6R expression by quantitative RT–PCR (qRT–PCR) is also shown. (B) Images and quantification of cytokine antibody arrays (measuring 42 different cytokines) incubated for 48 h with conditioned medium of either MCF-10A or MCF-10A/ErbB2* cells. Cytokines strongly secreted in MCF-10A/ErbB2* cells compared with MCF-10A cells are highlighted in the image by squares, and the same color code is used in the bar chart. The names of these secreted cytokines are shown above the bars. (C) Western blot analysis of STAT3 phosphorylation in MCF-10A cells when incubated with MCF-10A/ErbB2* conditioned medium for 1 h and inhibition of its activation when the cells are simultaneously treated with 50 µg/mL tocilizumab. (D) STAT3 phosphorylation after treatment of MCF-10A/ErbB2* and MCF-10A/ErbB2 with either 1 µM pan-Src inhibitor-I, 1 µM pan-JAK inhibitor-I, or 1 µM lapatinib for 8 h was measured by Western blot. (E) Determination of STAT3 phosphorylation in MCF-10A/ErbB2* cells after treatment with inhibitors of different JAK family members for 24 h. All inhibitors were used at 1 µM concentration except CEP33779, which was used at 5 µM. The table below indicates the specificity of each of the inhibitors. (F) Box plot representing the expression of IL-6 in primary breast cancers classified according to their estrogen receptor (ER) and HER2 status. For F and G, the data were obtained from the RPPA data set of the TCGA. Every dot represents a sample, and the dashed line indicates the slope of the correlation. (G) Correlation between STAT3 phosphorylation and IL-6 levels in each subtype of primary breast sample analyzed.
Phosphorylation and activation of STAT proteins is mostly mediated by JAK family members (Rawlings et al. 2004; Murray 2007). However, in cancer cells with constitutive growth factor receptor activation, it has also been shown that STAT proteins can also be activated by SRC kinases (Garcia et al. 2001). Thus, we used specific pan-inhibitors of JAK and SRC protein families as well as the dual EGFR/HER2 inhibitor lapatinib to identify the specific mechanism of STAT3 activation (Fig. 3D). These experiments showed that, in breast cancer cells, short-term inhibition of JAK proteins completely blocked STAT3 activation, while inhibition of SRC or HER2 did not have any effect. Note that we interrogated both MCF-10A overexpressing the oncogenic (MCF-10A/ErbB2*) and the wild-type HER2 (MCF-10A/ErbB2) gene because the mutant HER2 variant has been found to be resistant to lapatinib (Wang et al. 2006). Next, we used specific inhibitors of different JAK family members to identify which one of them was responsible for aberrant STAT3 activation, revealing JAK2 as its upstream activating kinase (Fig. 3E).
Remarkably, when we studied STAT3 phosphorylation levels (pY705) and IL-6 expression in primary breast tumors using the RPPA and mRNA expression data from the TCGA breast cancer data set (The Cancer Genome Atlas Network 2012), ER−/HER+ tumors presented both the highest levels of IL-6 expression (Fig. 3F) and the strongest correlation between IL-6 and STAT3 phosphorylation (Fig. 3G).
S100A8 and S100A9 are direct targets of STAT3, playing a critical role in HER2-mediated oncogenesis
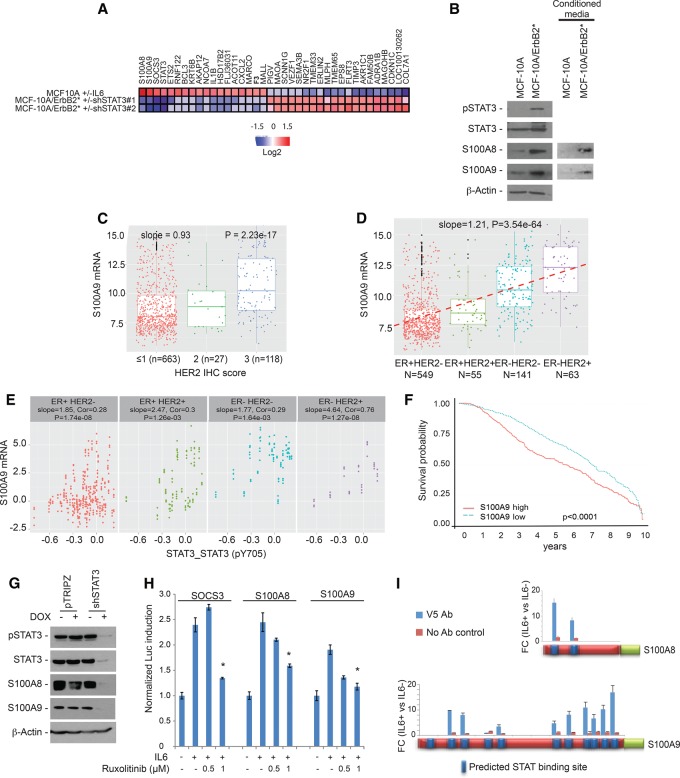
When activated, STAT3 works as a TF regulating target gene expression on the chromatin. In order to identify relevant targets of STAT3 in the ER−/HER2+ context, we compared gene expression profiles of MCF-10A cells following STAT3 activation (±IL-6 in the culture medium for 6 h) as well as of MCF-10A/ErbB2* cells following shRNA-mediated STAT3 silencing (Fig. 4A; Supplemental Fig. S4A). To identify targets that are differentially expressed following STAT3 activation, we selected genes that fulfill two criteria: (1) They are up/down-regulated more than twofold, with P < 0.05 in MCF-10A cells after IL-6 exposure, and (2) they showed an opposite trend in MCF-10A/ErbB2* cells following RNAi-mediated STAT3 silencing. As expected, STAT3 as well as SOCS3 were at the top of the list. Remarkably, other top genes included S100A8 and S100A9 (Fig. 4A; Supplemental Fig. S4B). High mRNA levels of S100A8/9 in our MCF-10A model led to high protein levels both in the cell extract and secreted in the medium (Fig. 4B).
Figure 4.
S100A8 and S100A9 are transcriptional targets of STAT3. (A) The heat map shows genes transcriptionally up-regulated in MCF-10A cells when STAT3 is activated by 50 µg/mL IL-6 for 6 h and down-regulated in MCF-10A/ErbB2* cells when STAT3 is silenced with two independent shRNAs. (B) Western blot analysis of S100A8 and S100A9 expression in MCF-10A and MCF-10AErbB2* whole extracts as well as in their 48-h conditioned medium. (C) Box plot representing the correlation between S100A9 mRNA expression and HER2 immunohistochemistry score in primary breast cancers. Data were obtained from a METABRIC data set. (D) Distribution of S100A9 mRNA expression in primary breast cancers according to tumor subtype as determined by ER and HER2 status. The slope of the correlation is shown by the dashed line and is indicated at the top. (E) Correlation between S100A9 mRNA levels and STAT3 phosphorylation in the primary breast cancer subtype. Data were obtained from the TCGA data set. (F) Ten-year Kaplan-Meier survival curve of breast cancer patients classified according to the S100A9 expression in the tumor. Data were obtained from the METABRIC data set. S100A9 levels above and under the median are considered high and low, respectively. (G) Analysis by Western blot of S100A8 and S100A9 levels when STAT3 expression is silenced by Dox-inducible shRNAs. (H) SOCS3, S100A8, and S100A9 promoters were cloned in a luciferase reporter vector. HEK293T cells were transfected with these constructs along with vectors expressing Renilla luciferase for normalization purpose. The bar graph represents the normalized levels of luciferase expression from each of these constructs when STAT3 was activated for 6 h with 50 µg/mL IL-6 in the presence of different doses of the dual JAK1/2 inhibitor ruxolitinib. (I) Determination of STAT3 binding to the S100A8 and S100A9 promoters was performed by chromatin immunoprecipitation (ChIP) assay using MCF-10A cells expressing V5-tagged STAT3. Blue squares in the promoter indicate putative STAT-binding sites, and the bars show the enrichment measured by qRT–PCR after STAT3-V5 immunoprecipitation of cells incubated for 30 min with 50 µg/mL IL-6. (*) Statistical significance < 0.05.
S100A8 and S100A9 are small cytoplasmic calcium-binding proteins that heterodimerize to form a complex (calprotectin) that is secreted into the intercellular space (Gebhardt et al. 2006; Ehrchen et al. 2009). Although they have been mainly studied in the context of the immune system, S100A8/9 have been found to be up-regulated in multiple solid tumors and associated with poor prognosis (Arai et al. 2008; Kawai et al. 2011; Grebhardt et al. 2014).
Next, we analyzed the METABRIC data to investigate whether overexpression of S100A8/9 was observed in ER−/HER2+ primary breast tumors. These studies revealed that mRNA levels of S100A8 and S100A9 are strongly correlated (r = 0.94), suggesting coregulation (Supplemental Fig. S4C). Furthermore, mRNA expression of S100A8/9 was strongly correlated with HER2 pathological status (Fig. 4C). Importantly, ER−/HER2+ tumors presented the highest S100A8/9 expression (Fig. 4D) as well as the highest correlation between STAT3 phosphorylation and S100A8/9 mRNA expression (the RPPA data set from the TCGA breast cancer study was used here) (Fig. 4E). Remarkably, high S100A8/9 expression was associated with lower breast cancer patient survival (Fig. 4F).
Taken together, these data suggest that expression of S100A8/9 in ER−/HER2+ tumor cells is intimately linked to aberrant STAT3 activation. To validate the dependency of S100A8 and S100A9 expression on STAT3 activity, we performed RNAi-mediated STAT3 silencing in MCF-10A/ErbB2* cells. As expected, STAT3 knockdown strongly reduced S100A8 and S100A9 expression (Fig. 4G). Both the S100A8 and S100A9 promoters present several predicted STAT-binding sequences. Thus, we cloned the S100A8, S100A9, and SOCS3 (positive control) promoters to drive expression of a luciferase reporter (Supplemental Fig. S4D) and evaluated reporter gene induction upon STAT3 activation. HEK293T cells transfected with the reporters showed strong induction of luciferase expression after addition of IL-6 to the medium (Fig. 4H; Supplemental Fig. S4E). Importantly, the effect was fully abrogated when STAT3 activation was prevented by the JAK1/2 inhibitor ruxolitinib (Fig. 4H; Mascarenhas and Hoffman 2012; Mesa et al. 2012). To further investigate whether regulation of S100A8 and S100A9 occurs by direct binding of STAT3 to their promoters, we performed chromatin immunoprecipitation (ChIP) assays. For this, we first engineered MCF-10A cells expressing V5-tagged STAT3. After checking that STAT3-V5 behaves identically to endogenous STAT3 (it is enriched in the nucleus after IL-6 exposure) (Supplemental Fig. S4F), STAT3-V5 was immunoprecipitated from MCF-10A cells cultured in the absence/presence of IL-6. Next, the presence of STAT3 in S100A8 and S100A9 promoters was evaluated by quantitative PCR (qPCR). These studies revealed a significant enrichment of STAT3 in both promoters upon IL-6-mediated STAT3 activation (Fig. 4I).
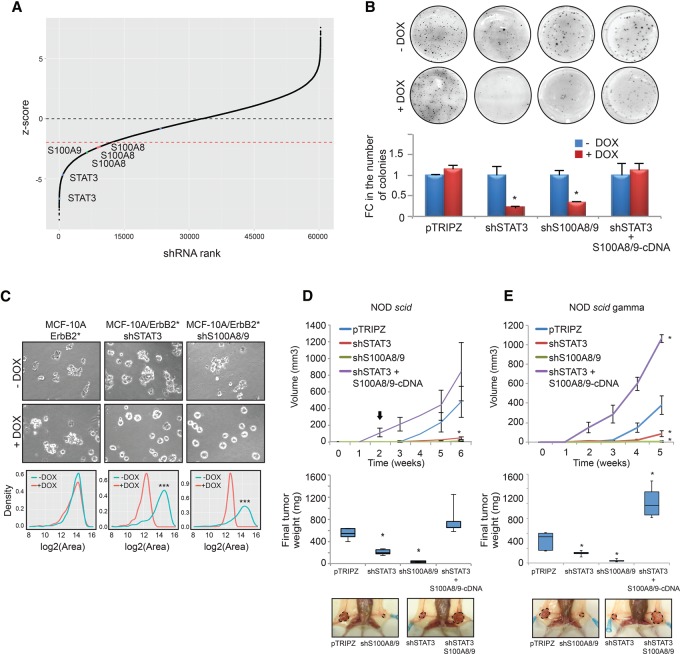
Since S100A8/9 are part of the transcriptional program up-regulated by STAT3 in ER−/HER2+ mammary tumors, we asked whether S100A8/9 are relevant for the essential role of STAT3 in these cancer cells. Remarkably, shRNAs targeting both S100A8 and S100A9 were also positive hits in our RNAi screen (Fig. 5A). To study the role of S100A8 and S100A9, we first engineered MCF-10A/ErbB2* cells to express shRNAs targeting both genes under inducible conditions (Supplemental Fig. S5A). We then studied how S100A8/9 silencing impacts the tumorigenic potential of these cells. S100A8/9 silencing phenocopied the lethal effect induced by STAT3 inhibition in vitro (Fig. 5B,C) and in vivo when MCF-10A/ErbB2* variants were orthotopically injected in NOD scid mice (Fig. 5D).
Figure 5.
S100A8 and S100A9 play a critical role in HER2-mediated oncogenesis. (A) Schematic representation of the rank of shRNAs according to their Z-scores in the screen. Positions of the shRNAs that target STAT3 (blue), S100A8 (orange), and S100A9 (green) are shown. (B) Colony formation assay in agar of MCF-10A/ErbB2* cells transduced with control vector (pTRIPZ), shRNA against STAT3 (shSTAT3), shRNAs against S100A8 and S100A9 (shS100A8/9), or shRNA against STAT3, where the expression of S100A8 and S100A9 has been restored experimentally by transduction of c-DNAs (shSTAT3 + S100A8/9-cDNA). All vectors expressing shRNAs were Dox-inducible. The chart bars represent the quantification of the number of colonies normalized to the pTRIPZ control. (C) Growth in ECM-Matrigel of cells from B. Quantification of the effect of gene silencing in the growth of the acini is provided in the graphs showing the size distribution of 50 acini. (***) P < 10−5. (D) The top panel shows the growth of tumors (average ± SD) when the different variants of MCF-10A/ErbB2* cells described in B were injected into the IMFP of NOD scid mice. n = 5. (E) The same experiments performed in D when the cells were injected into the IMFP of NOD scidγ mice. n = 6. The box plot represents the final weight of the dissected tumors, and a representative picture of each injected mouse is shown. The arrow indicates when 2 mg/mL Dox was added to the drinking water. (*) P < 0.05.
Next, we reasoned that, if STAT3-dependent lethality in MCF-10A/ErbB2* cells is mediated at least in part by loss of S100A8/9 expression, then re-expression of these genes should rescue the phenotype. To test this scenario, we first transduced MCF-10A/ErbB2*-expressing STAT3 targeting shRNAs with viral constructs expressing S100A8 and S100A9. As these viral constructs coexpress the cDNAs and fluorescent reporters, we were able to use FACS to separate the transduced cells in bins expressing different amounts of S100A8 and S100A9 based on fluorescence intensity (Supplemental Fig. S5B). Finally, we also selected sorted cells expressing S100A8 and S100A9 at the levels found in parental MCF-10A/ErbB2* cells (Supplemental Fig. S5B). When the tumorigenic potential of these cells was assayed, we found that re-expression of S100A8/9 restored the tumorigenic potential of MCF-10A/ErbB2* cells expressing STAT3 targeting shRNAs in vitro (Fig. 5B) and in vivo (Fig. 5D).
Because S100A8/9 are proinflammatory cytokines that influence the recruitment of immune cells (Gebhardt et al. 2006; Ehrchen et al. 2009), in order to eliminate, as much as possible, any contribution of the immune system in the growth of the tumor cells, we repeated the above studies using NOD scidγ mice (deficient in B and T cells, macrofages, NK cells, and complement). These experiments confirmed the importance of the autocrine stimulus mediated by S100A8/9 (Fig. 5E). Finally, we also asked whether up-regulation of S100A8/9 alone in nontransformed cells is able to transform them. Experimental up-regulation of S100A8 and S100A9 to the levels observed in HER2 transformed MCF-10A cells by using the viral cDNA constructs described above did not provide attachment-independent growth abilities to MCF-10A cells (Supplemental Fig. S5C).
S100A8 and S100A9 play a pivotal role in HER2-mediated oncogenesis by promoting key proliferative and survival signal transduction pathways
After demonstrating the critical role of S100A8/9 in HER2-mediated oncogenesis in our HR−/HER2+ model, we decided to investigate the specific mechanism. Exposure to S100A8/9 has been shown to activate canonical signal transduction pathways that are involved in tumorigenesis by increasing proliferation, resistance to stress, and protein synthesis (Gebhardt et al. 2006; Hermani et al. 2006; Ichikawa et al. 2011; Acharyya et al. 2012). As the activation of particular signaling is often context-dependent, we first decided to evaluate the activation of these pathways in our model after cells are exposed to recombinant S100A8/9 protein.
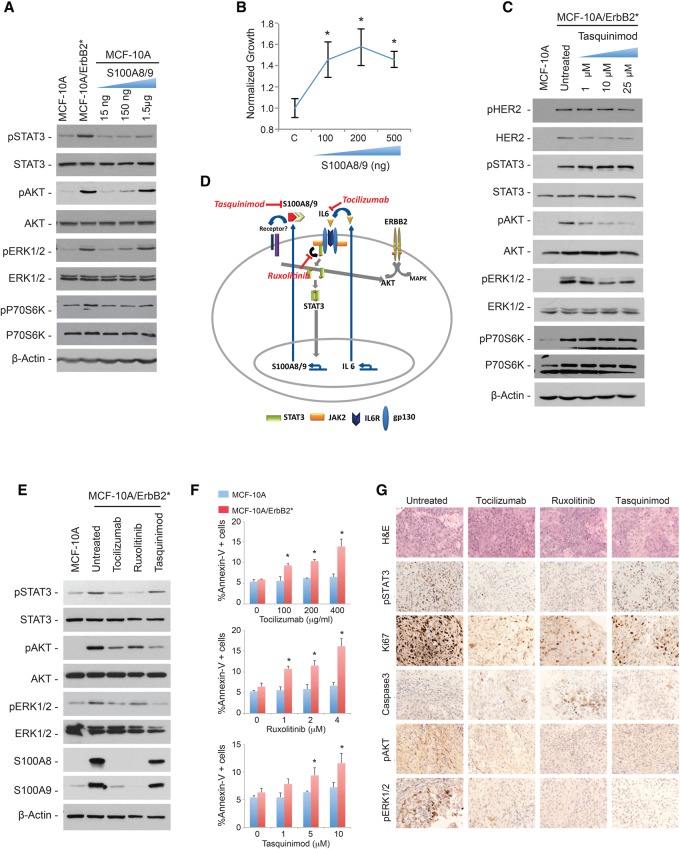
In order to expose cells to physiologic S100A8/9 protein levels, we first measured the amount of S100A8/9 proteins in the culture medium of MCF-10A/ErbB2* cells. Conditioned media with different numbers of cells growing exponentially for 48 h were concentrated, and the levels of S100A8/9 proteins were estimated by titrating recombinant protein through a standard curve (Supplemental Fig. S6A). This study revealed that ∼75–150 ng of S100A8 and S100A9 was present in the conditioned media per 1 × 105 cells (Supplemental Fig. S6A). When an equivalent amount of recombinant proteins was added to the culture medium of MCF-10A cells, we observed a robust increase in the phosphorylation levels (activation) of ERK1/2 and AKT and a minor effect on phosphor-P70S6K (Fig. 6A). Furthermore, MCF-10A cell proliferation increased (Fig. 6B). It is well established that activation of the above-mentioned pathways is directly achieved by EGFR/HER family members without the need for autocrine stimulation (Moasser 2007). Thus, it was important to investigate whether the activation mediated by the S100A8/9 autocrine mechanism is a way to amplify the signal and achieve the high levels observed in HER2+ cells or is simply redundant. To answer this question, we blocked S100A8/9 activity in MCF-10A/ErbB2* cells using the specific S100A9 inhibitor tasquinimod (Dalrymple et al. 2007, 2012). We found that, despite having the same HER2+ phosphorylation levels as untreated cells, blocking S100A8/9 significantly reduced the activation levels of AKT, while less pronounced effects were observed for ERK1/2 and P70S6K (Fig. 6C).
Figure 6.
S100A8 and S100A9 activate signal transduction pathways involved in tumorigenesis. (A) Western blot analysis of phosphorylation levels of AKT, ERK1/2, and p70S6K in MCF-10A cells incubated with increasing doses of recombinant S100A8 and S100A9 for 6 h. (B) Increase in the number of MCF-10A cells incubated with different doses of S100A8/9 for 48 h as measured with Cell Titer Glo. The fold change was normalized to untreated cells. (*) P < 0.05 (C) Phosphorylation levels of HER2, STAT3, AKT, ERK1/2, and p70S6K in MCF-10A/ErbB2* cells after treatment with increasing doses of the S100A9 inhibitor tasquinimod for 48 h. (D) Schematic representation of the double autocrine loop in HR−/HER2+ breast cancer cells. FDA-approved inhibitors against different components of the pathway are indicated in red. (E) The Western blot shows the phosphorylation levels of STAT3, AKT, and ERK1/2 as well as S100A8 and S100A9 levels after treatment with 50 µg/mL tocilizumab, 1 µM ruxolitinib, or 10 µM tasquinimod for 48 h. (F) Quantification of Annexin V+ cells by FACS in MCF-10A and MCF-10A/ErbB2* cells incubated with 50 µg/mL tocilizumab, 1 µM ruxolitinib, or 10 µM tasquinimod for 48 h. (*) P < 0.05 (G) Immunohistochemical analysis of pSTAT3, Ki67, Caspase3, pAKT, and pERK1/2 levels in MCF-10A/ErbB2* orthotopic tumors. Cells were injected into the IMFP of SCID mice and allowed to grow until they reached a volume of 0.3 cc, when the treatment was administered for 4 d as follows: 2 mg of tocilizumab per mouse, twice per week; 90 mg/kg ruxolitinib twice per day; or 10 mg/kg tasquinimod per day.
Our studies suggest the existence of a critical double autocrine stimulus in HR−/HER2+ breast cancer cells. The first stimulus is defined by the secretion of IL-6 to activate STAT3 through the canonical JAK pathway. Once activated, STAT3 induces up-regulation of the S1008/9 dimer, which is secreted and augments AKT activation and, to a lesser extent, ERK1/2 and P70S6K (Fig. 6D). Importantly, several compounds targeting key players of this double autocrine loop are already available in the clinic, providing exciting therapeutic opportunities (Fig. 6D; Supplemental Fig. S6B). Tocilizumab is a humanized antibody against the IL-6R. It is FDA-approved for the treatment of rheumatoid arthritis (RA) (Patel and Moreland 2010; Navarro-Millan et al. 2012). Ruxolitinib is a JAK1/2 dual inhibitor that has been recently FDA-approved to specifically treat patients with myelofibrosis (Mascarenhas and Hoffman 2012; Mesa et al. 2012). Tasquinimod binds to and inhibits the interactions of S100A9 and is actively being investigated for the treatment of solid tumors, mostly prostate cancers (Dalrymple et al. 2007, 2012).
Thus, we next studied the use of these inhibitors to reduce the levels of S100A8 and S100A9 proteins as well as the activation of the signaling cascades described above. As expected, treating MCF-10A/ErbB2* cells with tocilizumab or ruxolitinib strongly inhibited the expression of S100A8 and S100A9 (Fig. 6E), and each of the three inhibitors significantly attenuated phosphor-AKT levels. Remarkably, treatment with these inhibitors was associated with increased apoptosis in MCF-10A/ErbB2* cells compared with the parental MCF-10A line (Fig. 6F). Importantly, we observed comparable results in vivo when orthotopic models of MCF-10A/ErbB2* were treated with the same inhibitors (Fig. 6G; Supplemental Fig. S6C).
Inhibition of IL-6R/JAK2/S100A9 represents a novel target for patient-oriented therapies in HR−/HER2+ breast cancers
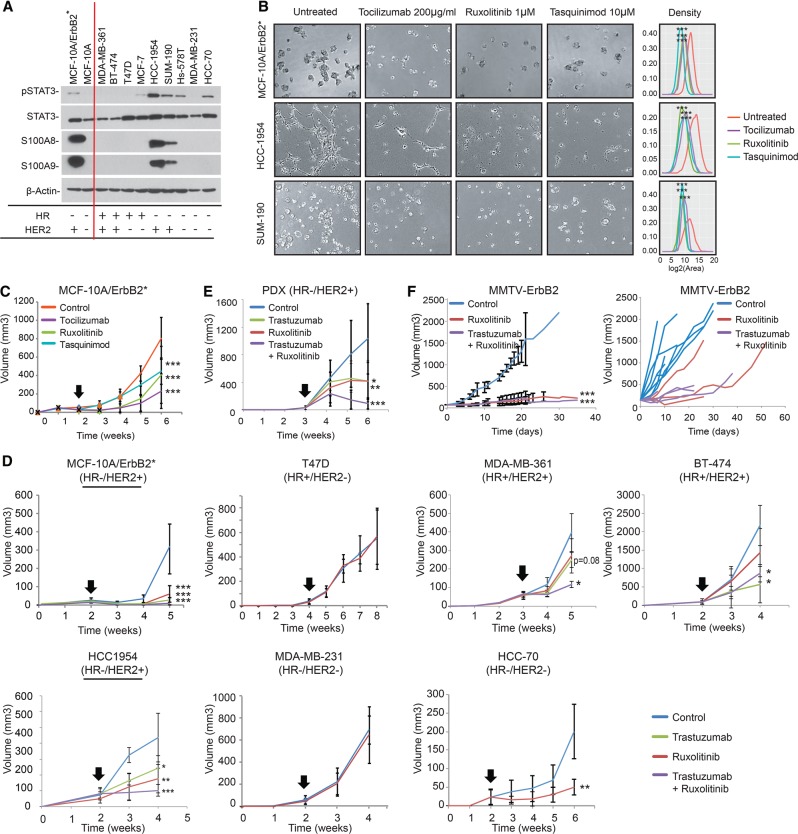
The studies described above suggest that inhibition of IL-6R/JAK2/S100A9 represents a novel, mechanistically motivated anti-cancer strategy in HR−/HER2+ breast cancers. In order to further investigate this possibility, we completed a series of in vitro and in vivo studies to evaluate tumor growth treatment with the above-described inhibitors. First, we selected a set of nine additional breast cancer cell lines, representing the four subtypes (HR+/HER2−, HR+/HER2+, HR−/HER2−, and HR−/HER2+) (Fig. 7A; Supplemental Fig. S7A). In these cells, we studied STAT3 phosphorylation as well as S100A8 and S100A9 protein expression. Consistent with our model, HR−/HER2+ cell lines presented both the highest phospho-STAT3 levels and the highest S100A8 and S100A9 expression (Fig. 7A). Furthermore, exposing HR−/HER2+ HCC-1954 cells to tocilizumab, ruxolitinib, and tasquinimod reduced the levels of p-STAT3, P-AKT, and S100A8/9, as seen in the MCF-10A/ErbB2* model (Supplemental Fig. S7B).
Figure 7.
Tocilizumab, ruxolitinib, and tasquinimod are efficient anti-cancer drugs in HR−/HER2+ breast cancer cells. (A) Western blot analysis of STAT3 phosphorylation as well as S100A8 and S100A9 levels in a panel of breast cancer cell lines. HR and HER2 status is indicated at the bottom. (B) Growth in ECM-Matrigel of HR−/HER2+ breast cancer models (MCF-10A/ErbB2*, HCC-1954, and SUM-190) when treated with inhibitors. Pictures were taken after 6 d of treatment with 200 µg/mL tocilizumab, 1 µM ruxolitinib, and 10 µM tasquinimod. A complete series of all 11 cell lines is shown in A, and additional drug doses are shown in Supplemental Figure S7. Quantification of the effect of drug treatment in the growth of the cells is provided in the graphs showing the size distribution of 50 acini. (***) P < 10−5. (C) Tumor growth of MCF-10A/ErbB2* cells injected into the IMFP of SCID mice. n = 6. Treatment was administrated as follows: 2 mg of tocilizumab per mouse, twice per week; 90 mg/kg ruxolitinib, twice per day; or 10 mg/kg tasquinimod per day. Representation of the growth curves generated by the injection of breast cancer cell lines (D) or a patient-derived xenograft (PDX) (E) into the IMFP of SCID mice. n = >6 for all animal experiments shown in D and E. (F) Three-month-old transgenic FVB-Tg(MMTV-Erbb2)NK1Mul/J female animals were mated and randomly assigned a treatment cohort when the absolute tumor mass reached 100 mm3. The left panel shows the growth of the tumor masses as absolute tumor volume per mouse (multifocal masses). The right panel shows the growth kinetics of individual tumors. Three individual animals were analyzed per cohort. For all panels, mice were treated with herceptin (one loading shot of 30 mg/kg followed by administration of 15 mg/kg twice per week), ruxolitinib (90 mg/kg, twice per day), or the combination of both. The black arrows indicate the point when the treatment was started (tumors reached 0.3 cc). (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
All of these cell models were then plated in Matrigel and individually treated with tocilizumab, ruxolitinib, and tasquinimod. As expected, growth of HR−/HER2+ cells was significantly affected by these treatments, while minor effects were observed in the remaining cells lines (Fig. 7B shows only HR−/HER2+ cell lines; Supplemental Fig. S7C,D shows the full set). Interestingly, in addition to the HR−/HER2+ models, HCC-70 was the only cell line that was strongly affected when treated with ruxolitinib. This was not surprising, as these cells have high levels of p-STAT3 (Fig. 7A). Finally, to transition our studies to an in vivo context, MCF-10A/ErbB2* cells were orthotopically transplanted into the mammary glands of NOD scid mice and individually treated with each of the three inhibitors. As seen in vitro, treatment with tocilizumab, ruxolitinib, and tasquinimod significantly reduced the tumorigenic potential of MCF-10A/ErbB2 cells in vivo (Fig. 7C; Supplemental Fig. S7E). To further complete our preclinical studies, seven different orthotopically injected cell lines in NOD scid mice representing the four subtypes, a patient-derived HR−/HER2+ xenograft, and a transgenic HER2+ breast cancer mouse model, FVB-Tg(MMTV-Erbb2)NK1Mul/J (http://jaxmice.jax.org/strain/005038.html), were treated with ruxolitinib. HER2+ models were also treated with the humanized anti-HER2 antibody trastuzumab (Lewis Phillips et al. 2008) and the combination of trastuzumab plus ruxolitinib. For these in vivo studies, ruxolitinib was selected over tasquinimod and tociluzumab because it is already FDA-approved for cancer treatment. These studies confirmed that response to ruxolitinib as a sole anti-cancer agent in all engrafted HR−/HER2+ cells (MCF-10A/ErbB2*, HCC-1954, and the patient-derived xenograft [PDX]) was statistically significant (Fig. 7D,E; Supplemental Fig. S7G,H). Interestingly, the HR+/HER2+ line MDA-MB-361 showed minimal response (not statistically significant) to individual treatment with ruxolitinib and trastuzumab, which became statistically significant when the animals were treated in combination. The enhanced anti-tumor effect when trastuzumab and ruxolitinib were used in combination was confirmed in both HR+/HER2+ and HR−/HER2+ models in vivo (Fig. 7D,E; Supplemental Fig. S7G,H). As seen in vitro, the HCC-70 model also responded to ruxolitinib treatment in vivo.
Transgenic FVB-Tg(MMTV-Erbb2)NK1Mul/J females developed multifocal tumors after pregnancy, and, consistent with the xenograft models, we observed that treatment with ruxolitinib reduced both the number of tumors formed and the growth of individual tumor masses (Fig. 7F).
Discussion
Breast cancer is not a homogeneous malignancy but rather a heterogeneous group of tumor diseases (Bertos and Park 2011). Hyperactivation of HER2 has been classically considered one of the determinants that define ∼20% of all breast cancers, and, consequently, HER2+ breast cancers have been managed in the clinic as a homogeneous group (Hynes and Lane 2005; Lemmon and Schlessinger 2010). However, emerging evidence has challenged this view, revealing important intrinsic differences associated with HR status (Perou et al. 2000; Paluch-Shimon et al. 2009; Sotiriou and Pusztai 2009; Vaz-Luis et al. 2012).
We and others have previously described screening strategies based on genetic (Silva et al. 2008; Marcotte et al. 2012) and small molecule (Gupta et al. 2009; Heiser et al. 2012) inhibitors aimed to identify the Achilles’ heel of tumor cells based on specific aspects of their biology. Similarly, we showed that analysis of regulatory networks can identify MR genes that are necessary and sufficient for tumor survival (Carro et al. 2010; Piovan et al. 2013; Aytes et al. 2014). Here, we describe the design and results of the first integrative analysis that combines both methodologies to identify novel therapeutic targets for HR−/HER2+ breast cancers. Our studies identified that HR−/HER2+ breast cancers robustly activate the IL-6–JAK2–STAT3–calprotectin cascade. Importantly, we also demonstrated that the tumorigenic potential of these cells is dependent on the activation of this autocrine cascade.
Activation of STAT3 in metastatic progression is well established and has been associated with myeloid cell recruitment (Chang et al. 2013), angiogenesis (Chang et al. 2013), epithelial–mesenchymal transition (Wendt et al. 2014), and migration (Barbieri et al. 2010a). However, its potential role in the maintenance of breast cancer cell homeostasis is not clear. While STAT3 is dispensable to initiate tumorigenesis in HER2-driven mouse mammary models (Barbieri et al. 2010b), blocking STAT3 signaling in some cancer cell models compromises their tumorigenicity (Marotta et al. 2011; Chang et al. 2013). Arguably, poor understanding of the molecular determinants that define STAT3 activation as an essential hub in some cells but not in others prevents us from considering therapies targeting its activation.
Here, we report that HR−/HER2+ breast cancers are especially sensitive to inhibition of STAT3 activation. Mechanistically, we identified the downstream targets of STAT3 (S100A8 and S100A9) as responsible, at least partially, for this sensitivity. S100A8 and S100A9 are calcium-binding proteins that heterodimerize, forming a complex called calprotectin that is secreted (Gebhardt et al. 2006; Ehrchen et al. 2009). While the role of S100A8/9 as proinflammatory mediators secreted by immune cells is well documented, S100A8/9 proteins are also found up-regulated in multiple solid tumors and associated with poor prognosis characteristics (Arai et al. 2008; Kawai et al. 2011; Grebhardt et al. 2014). Indeed, autocrine and paracrine exposure to calprotectin activates key signaling pathways involved in tumorigenesis (Gebhardt et al. 2006; Hermani et al. 2006; Ichikawa et al. 2011; Acharyya et al. 2012). Our data using immunocompromised mouse models lacking all major immune cells (NOD scidγ) reveal that, in the context of HR−/HER2+ breast cancers, S100A8/9 secreted by the tumor cells is an important mediator of tumorigenesis that increases the phosphorylation of AKT and, to a lesser extent, ERK.
How can the above-mentioned observations explain the sensitivity of HR−/HER2+ breast cancer cells to inhibition of the IL-6–JAK2–STAT3–calprotectin cascade? Aberrant AKT activity is a key event commonly seen in human cancers that positively impacts most cancer hallmarks, including cell cycle, survival, metabolism, motility, genomic instability, angiogenesis, and inflammatory cell recruitment (Testa and Tsichlis 2005; Fruman and Rommel 2014). AKT is directly stimulated by activation of growth factor receptors, including HER2, through canonical PI3K–PIP3 signals. Additionally, cross-talk with other pathways and feedback loops modulate the final activity of AKT (Testa and Tsichlis 2005; Fruman and Rommel 2014). Our results suggest that overexpression and secretion of S100A8 and S100A9, as detected in HR−/HER2+ cells, also contribute to aberrant AKT activity. It is thus plausible that inhibiting S100A8/9 secretion may reduce AKT activity (despite an otherwise unchanged stimulus from oncogenic HER2) to a point that becomes critical for these cancer cells. Although several receptors for S100A8/9 have been identified (RAGE [Turovskaya et al. 2008], TLR4 [Kallberg et al. 2012], and EMMPRIM [Hibino et al. 2013]), how the signal is transmited to AKT is not characterized. It will be important to dissect the signaling pathways that connect S100A8/9 with AKT, as additional therapeutic alternatives may emerge.
An additional consideration regarding the sensitivity of HR−/HER2+ cells is that STAT3 has been found activated in stem cell-like breast cancer cells, and its inhibition was shown to reduce their viability (Marotta et al. 2011; Chung et al. 2014). Thus, the anti-tumor response observed may also be a reflex of the inhibition of the stem cells inside HR−/HER2+ cancers. Although HR−/HER2+ cancer cells were the most sensitive to the inhibition of the IL-6–JAK2–STAT3–calprotectin axis, we observed that STAT3 blockade with ruxolitinib also affects the growth of a HR+/HER2+ model—especially when combined with anti-HER2 therapy—as well as some breast cancer cells with high levels of p-STAT3 (see Fig. 7D; Supplemental Fig. 7G). Remarkably, the link between HER2, STAT3, and stem cell-like cells has also been described in HR+ breast cancers, which could explain this effect (Marotta et al. 2011; Chung et al. 2014). Overall, the studies described here unveil the IL-6–JAK2–STAT3–calprotectin cascade as an Achilles’ heel of HR−/HER2+ tumors. Importantly, our preclinical studies demonstrating anti-cancer activity of FDA-approved drugs (ruxolitinib and tociluzumab) as well as of a compound that is currently being evaluated in clinical trials (tasquinimod) open new avenues for a rapid transition of our findings to the clinic. Indeed, our data serve as the basis for investigating the combination of ruxolitinib and trastuzumab in a phase I/II, multicenter trial in patients with metastatic breast cancer that has progressed on prior HER2 targeted therapy (ClinicalTrials.gov identifier NCT02066532). If a clinical benefit is identified, inhibition of the IL-6–JAK–STAT3–calprotectin axis by ruxolitinib would represent a novel, non-chemotherapy-containing, orthogonal anti-cancer strategy for HER2+ breast cancers.
Materials and methods
Pooled shRNA screening
MCF-10A and MCF-10A/ErbB2* cells were infected with a pool of virus generated from the Open Biosystem GIPZ lentiviral human shRNA library that comprises 58,493 shRNA targeting 18,651 genes. After infection, cells were cultured in triplicate in standard plastic plates for 10 doubling times or embedded in ECM-Matrigel for 1 wk. In addition, MCF-10A/ErbB2* cells were injected in triplicate into the intramammary fat pad (IMFP) of immunocompromised mice to perform the in vivo screening. Experimental details are described in Rodriguez-Barrueco et al. (2013). Data analysis details are described in Yu et al. (2013).
Data-driven network reconstruction of breast cancer
We used a data-driven approach, ARACNe, to reconstruct a breast cancer interactome from 359 TCGA breast cancer gene expression profiles. We then applied ARACNe against 1597 probes corresponding to 780 TFs to establish a TF-centered interactome and against 6434 probes for 2453 signaling molecule genes to construct a SP-focused network. The parameters of the algorithm were configured as follows: P-value threshold, P = 1 × 10−7; DPI tolerance, e = 0; and number of bootstraps, NB = 100. We used the adaptive partitioning algorithm for mutual information estimation.
Gene expression arrays
For gene expression arrays, MCF-10A and MCF-10A/ErbB2* RNAs were extracted using RNeasy extraction kits and labeled using the low-input QuickAmp labeling kit (Agilent, no. 5190-2331) following the manufacturer's instructions. Finally, labeled RNAs were hybridized on an Illumina HT12v3 microarray. The experiment was performed with an n = 6.
MCF-10A cells were treated with 50 µg/mL IL6 for 1 h. In parallel, MCF-10A/ErbB2* infected with inducible shRNAs against STAT3 were incubated with 100 ng/mL Dox for 5 d. RNA from each condition was extracted with RNeasy extraction kit, labeled, and hybridized on a human GE 4x44K version 2 microarray kit (Agilent, no. G4845A).
Gene expression data have been uploaded, and the Gene Expression Omnibus accession number assigned is GSE62251.
MR analysis
We interrogated the TCGA breast cancer interactomes and applied MARINa to identify key master transcriptional or signaling drivers for MCF-10A/ErbB2* cells or HR−/HER2+ breast cancer samples in TCGA or METABRIC data. For the gene set enrichment analysis method in MARINa, we applied the “maxmean” statistic to score the enrichment of the gene set and used a sample permutation to build the null distribution for statistical significance.
Viral production and infection
Lentiviral production was achieved by transfecting phoenix packaging cells with jet-PEI (Polyplus, #101-10N) in combination with the lentiviral plasmids (shRNA listed below, pLOC_S100A8, and pLX304-Blast-V5_STAT3) and the pCMV-dR8.91 and pMD.G helper plasmids at a ratio of 2:1:1, respectively (Rodriguez-Barrueco et al. 2013). The same conditions were employed to produce retroviral particles by combining the retroviral plasmids (pBABE-HERYVMA and pLPCX-RFP_S100A9) with the retroviral helper plasmids Psi and VSVg. A detailed protocol is in the Supplemental Material as well as precious publications (Rodriguez-Barrueco et al. 2013).
S0CS3, S100A8, and S100A9 promoter cloning and luciferase assay
To measure luciferase activity, phoenix cells were plated at 70% confluence in 96-well plates. Twenty-four hours later, cells were transfected with 50 ng of pGL3 constructs containing the promoter sequences in combination with a Renilla normalization control using jet-PEI transfection reagent. After 24 h, relative luciferase units (RLU) were measured using the Dual-Glo Luciferase assay system (Promega, #E2949).
Soft agar colony formation assay
Cells were plated in semisolid medium as follows: 35-mm plates were layered with 0.6% agar (Spectrum Chemical, AG110) and medium, and 5.0 × 103 cells per milliliter were seeded in triplicate in the second layer of 0.3% agar and medium and cultured in the appropriate growth medium. Colonies were stained with 1 mg/mL MTT (Sigma, M2128) after 3 wk of incubation, photographed, and counted, and the average ± standard deviation was represented.
3D basement membrane cultures
Wells in 24-well low-attachment plates (Corning, no. 3473) were precoated with a layer of 200 µL of 100% ECM-Matrigel per well (BD Biosciences, no. 354230) and allowed to gel at 37°C. Cells were trypsinized and diluted at densities of 1 × 105 or 2 × 105 cells per milliliter in assay medium (DMEM/F12 supplemented with 5% HS, 0.5 µg/mL hydrocortisone, 100 ng/mL cholera toxin, 10 µg/mL insulin, 4 ng/mL EGF, 5% ECM-Matrigel). Fresh assay medium containing the drugs at the indicated concentrations was added every day. Pictures were taken after 6 d of treatment.
Mice
Animal maintenance and experiments were performed in accordance with the animal care guidelines and protocols approved by the Columbia University animal care unit. Eight-week-old female NOD.CB17-Prkdcs SCID mice (Harlan) were injected with 5 × 106 cells resuspended in Matrigel (BD Biosciences):normal growth medium (1:1) into the fat pad mammary gland. Dox was added to drinking water at a final concentration of 2.0 mg/mL. Tumor growth was monitored twice a week with calipers at the site of injection. Animals were sacrificed when tumor size reached 1.5-cm diameter.
ChIP
MCF-10A and MCF-10A/STAT3-V5 cells (untreated cells and cells treated with 50 µg/mL IL6 for 30 min) were grown to 80%–90% confluency in 150-mm culture dishes. After washing with PBS, cells were fixed and cross-linked using a truChIP High Cell chromatin shearing kit with SDS shearing buffer (Covaris, no. 010128) following the manufacturer's instructions.
The next day, anti-V5-conjugated beads were resuspended in 1 mL of blocking buffer. Nuclear extracts (100 µL) were added to the bead solution and incubated overnight at 4°C on a rotator. After 12 h, beads were collected, and DNA was obtained by standard protocols (detailed in the Supplemental Material).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant R01/EUREKA R01CA153233 and the American Association for Cancer Research-Stand up to Cancer award (J.M.S.) and the National Cancer Center (R.R.-B.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.262642.115.
References
- Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N, Seshan VE, et al. 2012. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 150: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai K, Takano S, Teratani T, Ito Y, Yamada T, Nozawa R. 2008. S100A8 and S100A9 overexpression is associated with poor pathological parameters in invasive ductal carcinoma of the breast. Curr Cancer Drug Targets 8: 243–252. [DOI] [PubMed] [Google Scholar]
- Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, Eastham JA, Gopalan A, Pienta KJ, Shen MM, et al. 2014. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell 25: 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I, Pensa S, Pannellini T, Quaglino E, Maritano D, Demaria M, Voster A, Turkson J, Cavallo F, Watson CJ, et al. 2010a. Constitutively active Stat3 enhances neu-mediated migration and metastasis in mammary tumors via upregulation of Cten. Cancer Res 70: 2558–2567. [DOI] [PubMed] [Google Scholar]
- Barbieri I, Quaglino E, Maritano D, Pannellini T, Riera L, Cavallo F, Forni G, Musiani P, Chiarle R, Poli V. 2010b. Stat3 is required for anchorage-independent growth and metastasis but not for mammary tumor development downstream of the ErbB-2 oncogene. Mol Carcinog 49: 114–120. [DOI] [PubMed] [Google Scholar]
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. 2005. Reverse engineering of regulatory networks in human B cells. Nat Genet 37: 382–390. [DOI] [PubMed] [Google Scholar]
- Berclaz G, Altermatt HJ, Rohrbach V, Siragusa A, Dreher E, Smith PD. 2001. EGFR dependent expression of STAT3 (but not STAT1) in breast cancer. Int J Oncol 19: 1155–1160. [DOI] [PubMed] [Google Scholar]
- Berishaj M, Gao SP, Ahmed S, Leslie K, Al-Ahmadie H, Gerald WL, Bornmann W, Bromberg JF. 2007. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res 9: R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertos NR, Park M. 2011. Breast cancer—one term, many entities? J Clin Invest 121: 3789–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2012. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. 2010. The transcriptional network for mesenchymal transformation of brain tumours. Nature 463: 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Q, Bournazou E, Sansone P, Berishaj M, Gao SP, Daly L, Wels J, Theilen T, Granitto S, Zhang X, et al. 2013. The IL-6/JAK/Stat3 feed-forward loop drives tumorigenesis and metastasis. Neoplasia 15: 848–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SS, Giehl N, Wu Y, Vadgama JV. 2014. STAT3 activation in HER2-overexpressing breast cancer promotes epithelial–mesenchymal transition and cancer stem cell traits. Int J Oncol 44: 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Yarden Y. 2006. EGF–ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 7: 505–516. [DOI] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalrymple SL, Becker RE, Isaacs JT. 2007. The quinoline-3-carboxamide anti-angiogenic agent, tasquinimod, enhances the anti-prostate cancer efficacy of androgen ablation and taxotere without effecting serum PSA directly in human xenografts. Prostate 67: 790–797. [DOI] [PubMed] [Google Scholar]
- Dalrymple SL, Becker RE, Zhou H, DeWeese TL, Isaacs JT. 2012. Tasquinimod prevents the angiogenic rebound induced by fractionated radiation resulting in an enhanced therapeutic response of prostate cancer xenografts. Prostate 72: 638–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE. 2005. Validating Stat3 in cancer therapy. Nat Med 11: 595–596. [DOI] [PubMed] [Google Scholar]
- Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. 2002. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell 111: 29–40. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30: 256–268. [DOI] [PubMed] [Google Scholar]
- Ehrchen JM, Sunderkotter C, Foell D, Vogl T, Roth J. 2009. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol 86: 557–566. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Rommel C. 2014. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov 13: 140–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muro-Cacho CA, Cox CE, Falcone R, Fairclough R, Parsons S, et al. 2001. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene 20: 2499–2513. [DOI] [PubMed] [Google Scholar]
- Gebhardt C, Nemeth J, Angel P, Hess J. 2006. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol 72: 1622–1631. [DOI] [PubMed] [Google Scholar]
- Grebhardt S, Muller-Decker K, Bestvater F, Hershfinkel M, Mayer D. 2014. Impact of S100A8/A9 expression on prostate cancer progression in vitro and in vivo. J Cell Physiol 229: 661–671. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. 2009. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138: 645–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman ZC, Yang XY, Glass O, Lei G, Osada T, Dave SS, Morse MA, Clay TM, Lyerly HK. 2011. HER2 overexpression elicits a proinflammatory IL-6 autocrine signaling loop that is critical for tumorigenesis. Cancer Res 71: 4380–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, Gibb WJ, Wang NJ, Ziyad S, Tong F, et al. 2012. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci 109: 2724–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermani A, De Servi B, Medunjanin S, Tessier PA, Mayer D. 2006. S100A8 and S100A9 activate MAP kinase and NF-κB signaling pathways and trigger translocation of RAGE in human prostate cancer cells. Exp Cell Res 312: 184–197. [DOI] [PubMed] [Google Scholar]
- Hibino T, Sakaguchi M, Miyamoto S, Yamamoto M, Motoyama A, Hosoi J, Shimokata T, Ito T, Tsuboi R, Huh NH. 2013. S100A9 is a novel ligand of EMMPRIN that promotes melanoma metastasis. Cancer Res 73: 172–183. [DOI] [PubMed] [Google Scholar]
- Hurley J, Doliny P, Reis I, Silva O, Gomez-Fernandez C, Velez P, Pauletti G, Powell JE, Pegram MD, Slamon DJ. 2006. Docetaxel, cisplatin, and trastuzumab as primary systemic therapy for human epidermal growth factor receptor 2-positive locally advanced breast cancer. J Clin Oncol 24: 1831–1838. [DOI] [PubMed] [Google Scholar]
- Hynes NE, Lane HA. 2005. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 5: 341–354. [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Williams R, Wang L, Vogl T, Srikrishna G. 2011. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol Cancer Res 9: 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg E, Vogl T, Liberg D, Olsson A, Bjork P, Wikstrom P, Bergh A, Roth J, Ivars F, Leanderson T. 2012. S100A9 interaction with TLR4 promotes tumor growth. PLoS One 7: e34207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman B, Mackey JR, Clemens MR, Bapsy PP, Vaid A, Wardley A, Tjulandin S, Jahn M, Lehle M, Feyereislova A, et al. 2009. Trastuzumab plus anastrozole versus anastrozole alone for the treatment of postmenopausal women with human epidermal growth factor receptor 2-positive, hormone receptor-positive metastatic breast cancer: results from the randomized phase III TAnDEM study. J Clin Oncol 27: 5529–5537. [DOI] [PubMed] [Google Scholar]
- Kawai H, Minamiya Y, Takahashi N. 2011. Prognostic impact of S100A9 overexpression in non-small cell lung cancer. Tumour Biol 32: 641–646. [DOI] [PubMed] [Google Scholar]
- Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, Wang K, Sumazin P, Kustagi M, Bisikirska BC, et al. 2010. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol 6: 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. 2010. Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis Phillips GD, Li G, Dugger DL, Crocker LM, Parsons KL, Mai E, Blattler WA, Lambert JM, Chari RV, Lutz RJ, et al. 2008. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res 68: 9280–9290. [DOI] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. 2008. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci 105: 20380–20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte R, Brown KR, Suarez F, Sayad A, Karamboulas K, Krzyzanowski PM, Sircoulomb F, Medrano M, Fedyshyn Y, Koh JL, et al. 2012. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov 2: 172–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. 2006. ARACNe: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ, Choudhury SA, Maruyama R, et al. 2011. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24− stem cell-like breast cancer cells in human tumors. J Clin Invest 121: 2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas J, Hoffman R. 2012. Ruxolitinib: the first FDA approved therapy for the treatment of myelofibrosis. Clin Cancer Res 18: 3008–3014. [DOI] [PubMed] [Google Scholar]
- Mesa RA, Yasothan U, Kirkpatrick P. 2012. Ruxolitinib. Nat Rev Drug Discov 11: 103–104. [DOI] [PubMed] [Google Scholar]
- Moasser MM. 2007. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 26: 6469–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ. 2007. The JAK–STAT signaling pathway: input and output integration. J Immunol 178: 2623–2629. [DOI] [PubMed] [Google Scholar]
- Navarro-Millan I, Singh JA, Curtis JR. 2012. Systematic review of tocilizumab for rheumatoid arthritis: a new biologic agent targeting the interleukin-6 receptor. Clin Ther 34: 788–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch-Shimon S, Ben-Baruch N, Wolf I, Zach L, Kopolovic J, Kruglikova A, Modiano T, Yosepovich A, Catane R, Kaufman B. 2009. Hormone receptor expression is associated with a unique pattern of metastatic spread and increased survival among HER2-overexpressing breast cancer patients. Am J Clin Oncol 32: 504–508. [DOI] [PubMed] [Google Scholar]
- Patel AM, Moreland LW. 2010. Interleukin-6 inhibition for treatment of rheumatoid arthritis: a review of tocilizumab therapy. Drug Des Devel Ther 4: 263–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. 2000. Molecular portraits of human breast tumours. Nature 406: 747–752. [DOI] [PubMed] [Google Scholar]
- Piovan E, Yu J, Tosello V, Herranz D, Ambesi-Impiombato A, Da Silva AC, Sanchez-Martin M, Perez-Garcia A, Rigo I, Castillo M, et al. 2013. Direct reversal of glucocorticoid resistance by AKT inhibition in acute lymphoblastic leukemia. Cancer Cell 24: 766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings JS, Rosler KM, Harrison DA. 2004. The JAK/STAT signaling pathway. J Cell Sci 117: 1281–1283. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Barrueco R, Marshall N, Silva J. 2013. Pooled shRNA screenings: experimental approach. Methods Mol Biol 980: 353–370. [DOI] [PubMed] [Google Scholar]
- Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al. 2008. Cancer proliferation gene discovery through functional genomics. Science 319: 620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz N, Marenstein DR, De Angelis DA, Wang WQ, Nelander S, Jacobsen A, Marks DS, Massague J, Sander C. 2011. Off-target effects dominate a large-scale RNAi screen for modulators of the TGF-β pathway and reveal microRNA regulation of TGFBR2. Silence 2: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, Elledge SJ, Hannon GJ, Chang K. 2008. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science 319: 617–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C, Pusztai L. 2009. Gene-expression signatures in breast cancer. N Engl J Med 360: 790–800. [DOI] [PubMed] [Google Scholar]
- Soule HD, Maloney TM, Wolman SR, Peterson WD Jr, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. 1990. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res 50: 6075–6086. [PubMed] [Google Scholar]
- Tagliabue E, Balsari A, Campiglio M, Pupa SM. 2010. HER2 as a target for breast cancer therapy. Expert Opin Biol Ther 10: 711–724. [DOI] [PubMed] [Google Scholar]
- Testa JR, Tsichlis PN. 2005. AKT signaling in normal and malignant cells. Oncogene 24: 7391–7393. [DOI] [PubMed] [Google Scholar]
- Turovskaya O, Foell D, Sinha P, Vogl T, Newlin R, Nayak J, Nguyen M, Olsson A, Nawroth PP, Bierhaus A, et al. 2008. RAGE, carboxylated glycans and S100A8/A9 play essential roles in colitis-associated carcinogenesis. Carcinogenesis 29: 2035–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz-Luis I, Ottesen RA, Hughes ME, Marcom PK, Moy B, Rugo HS, Theriault RL, Wilson J, Niland JC, Weeks JC, et al. 2012. Impact of hormone receptor status on patterns of recurrence and clinical outcomes among patients with human epidermal growth factor-2-positive breast cancer in the National Comprehensive Cancer Network: a prospective cohort study. Breast Cancer Res 14: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SE, Narasanna A, Perez-Torres M, Xiang B, Wu FY, Yang S, Carpenter G, Gazdar AF, Muthuswamy SK, Arteaga CL. 2006. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell 10: 25–38. [DOI] [PubMed] [Google Scholar]
- Weerasinghe P, Garcia GE, Zhu Q, Yuan P, Feng L, Mao L, Jing N. 2007. Inhibition of Stat3 activation and tumor growth suppression of non-small cell lung cancer by G-quartet oligonucleotides. Int J Oncol 31: 129–136. [PubMed] [Google Scholar]
- Wendt MK, Balanis N, Carlin CR, Schiemann WP. 2014. STAT3 and epithelial–mesenchymal transitions in carcinomas. JAKSTAT 3: e28975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormald S, Hilton DJ. 2004. Inhibitors of cytokine signal transduction. J Biol Chem 279: 821–824. [DOI] [PubMed] [Google Scholar]
- Yu J, Putcha P, Califano A, Silva JM. 2013. Pooled shRNA screenings: computational analysis. Methods Mol Biol 980: 371–384. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.