Abstract
The pre-mRNA splicing reaction of eukaryotic cells has to be carried out extremely accurately, as failure to recognize the splice sites correctly causes serious disease. The small subunit of the U2AF heterodimer is essential for the determination of 3′ splice sites in pre-mRNA splicing, and several single-residue mutations of the U2AF small subunit cause severe disorders such as myelodysplastic syndromes. However, the mechanism of RNA recognition is poorly understood. Here we solved the crystal structure of the U2AF small subunit (U2AF23) from fission yeast, consisting of an RNA recognition motif (RRM) domain flanked by two conserved CCCH-type zinc fingers (ZFs). The two ZFs are positioned side by side on the β sheet of the RRM domain. Further mutational analysis revealed that the ZFs bind cooperatively to the target RNA sequence, but the RRM domain acts simply as a scaffold to organize the ZFs and does not itself contact the RNA directly. This completely novel and unexpected mode of RNA-binding mechanism by the U2AF small subunit sheds light on splicing errors caused by mutations of this highly conserved protein.
Keywords: RNA splicing, 3′ splice site, U2AF small subunit
In eukaryotic cells, protein-coding genes are first transcribed into their precursor mRNA (pre-mRNA) transcripts, which have nontranslated intervening sequences called introns (Will and Lührmann 2011). The introns are excised from pre-mRNA by splicing reactions carried out in the nucleus, yielding mature mRNA ready to be exported to the cell cytoplasm. Introns possess consensus sequences that define the 5′ splice site, branch site, and 3′ splice site (Will and Lührmann 2011), which are recognized by the U1 snRNP, SF1/BBP protein, and U2AF heterodimer, respectively (Zamore and Green 1989; Jurica and Moore 2003; Wahl et al. 2009). The binding of the U2AF heterodimer to the 3′ splice site, which contains a well-conserved AG dinucleotide sequence, recruits U2 anRNP at the branch site, replacing the SF1/BBP protein, and the interactions between U1 and U2 snRNPs evoke the splicing reaction in an ATP-dependent manner (Will and Lührmann 2011). These factors play a crucial role in determining the splice sites, which has to be carried out extremely accurately to avoid generating aberrant mRNA. Misdirected splicing reactions are known to cause several serious human diseases (Yoshida et al. 2011; Imielinski et al. 2012; Brooks et al. 2014; Shao et al. 2014).
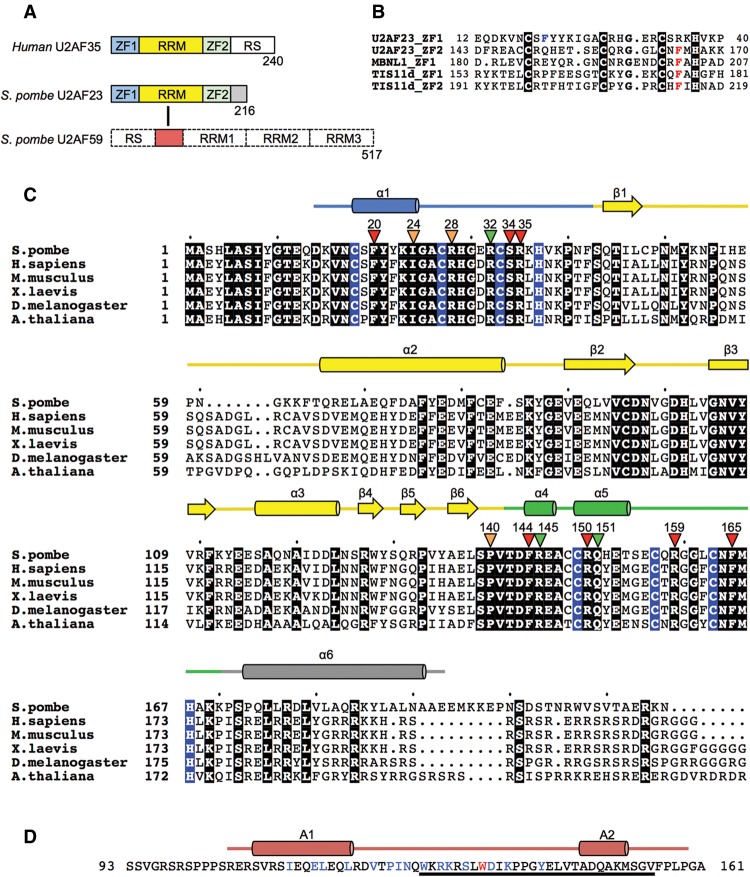
The U2AF heterodimer consists of large and small subunits (U2AF65 and U2AF35 in humans and U2AF59 and U2AF23 in fission yeast, respectively) (Zamore and Green 1989). The large subunit possesses three consecutive RNA recognition motifs (RRMs), and the small subunit has one RRM domain flanked by two CCCH-type zinc fingers (ZFs) (Fig. 1A; Mollet et al. 2006). A 28-residue fragment just before RRM1 in the large subunit (the corresponding region in the yeast U2AF59 is indicated by an underline in Fig. 1D) was reported to interact with the RRM domain of the small subunit (Kielkopf et al. 2001).
Figure 1.
Domain architecture and amino acid sequence of the U2AF small subunit. (A) Representative diagram of the domain architecture of the human and Schizosaccharomyces pombe U2AF small subunit. The RRM, ZF1, and ZF2 are colored yellow, blue, and green, respectively. The interaction region of U2AF59 with U2AF23 is shown as a solid dark-red box. (B) Multiple-sequence alignment of the CCCH-type ZFs. Residues blocked in black coordinate the zinc atom. Nearly completely conserved aromatic amino acid residues (Phe or Tyr) are shown in red. A residue that forms a hydrophobic convex in ZF1 of U2AF23 is shown in blue. (C) Multiple-sequence alignment of the U2AF small subunits. Numbering is based on the S. pombe U2AF23. Residues blocked in black are conserved among these orthologous species, and residues blocked in blue coordinate the zinc atom. The secondary structure is indicated with pink bars for α helices and with blue arrows for β strands. Arrows on the sequence indicate the residues mutated in the present study to examine their RNA-binding activity. (Green) No effect; (orange) 10 times less than that of wild type; (red) abolished. Amino acid residues composing ZF1, RRM, and ZF2 are boxed by the solid lines with the same colors as in A, respectively. The GeneBank accession numbers are as follows: Homo sapiens, Q01081; Mus musculus, Q9D883; Xenopus laevis, Q32NM8; Drosophila melanogaster, Q94535; Arabidopsis thaliana, Q9FMY5; S. pombe, Q09176. (D) Amino acid sequence of the peptide fragment of U2AF59 used for the present study. The ssecondary structure is indicated with red bars for α helices. The residue that is involved in “reciprocal Trp recognition” (Trp135) is colored in red. Residues that interact with U2AF23 are shown in blue.
By biochemical and structural study, it has been revealed that the human U2AF large subunit binds to the consecutive polypyrimidine tract (PPT) adjacent to the AG dinucleotide of the 3′ splice site through its two RRM domains (RRM1 and RRM2) (Zamore et al. 1992; Mollet et al. 2006; Sickmier et al. 2006). On the other hand, several lines of biochemical evidence indicate that the small subunit enhances the ability of the U2AF heterodimer to bind tightly to 3′ splice sites in substrate RNA (Zuo and Maniatis 1996; Rudner et al. 1998). U2AF35 can be chemically cross-linked to an AG dinucleotide sequence but not GG or UG (Merendino et al. 1999; Wu et al. 1999). Since the two CCCH-type ZFs and RRM of the U2AF small subunit are highly conserved among eukaryote species (Fig. 1C), it is widely considered that these domains play a crucial role in the RNA recognition mode of U2AF. However, the particular role of each domain remains controversial in the absence of any molecular model of the protein.
Canonical RRM domains bind to RNA directly (hence the name), and it has been reported that the U2AF35 RRM is sufficient to activate splicing of the mouse immunoglobulin M pre-mRNA (Guth et al. 2001). A minimal U2AF complex consisting of the RRM domain of U2AF35 and the above-mentioned 28-residue fragment of U2AF65 bound with micromolar affinity (Kd = 6 μM) to a 24-nucleotide RNA sequence containing both a PPT and the 3′ splice site AGGU nucleotides (Kielkopf et al. 2001). However, there are few perturbations in the NMR spectrum of the U2AF35 RRM domain upon addition of cognate RNA (Soares et al. 2006), which raises the question of the true role of the RRM of the U2AF small subunit in 3′ splice site binding.
CCCH-type ZFs are found in many RNA-binding proteins, such as TIS11d, and are often associated with RNA-binding ability. Actually, substitutions of the zinc-coordinated cysteine residues of either ZF in U2AF23 are lethal (Webb and Wise 2004), and many other single-residue mutations in these ZFs cause abnormal RNA splicing, leading to human pathogenesis (Yoshida et al. 2011; Imielinski et al. 2012; Brooks et al. 2014; Shao et al. 2014). However, the first ZF (ZF1) of the U2AF small subunit lacks the essential aromatic amino acid residues found in canonical RNA-binding CCCH-type ZFs (Fig. 1B). Furthermore, the two ZFs of the U2AF small subunit adopt a domain architecture distinctly different from that of previously studied CCCH-type ZFs with RNA-binding activity, all of which are tandemly linked in the amino acid sequence.
In order to understand the mechanism of 3′ splice site selection by the U2AF heterodimer and resolve the conflicting results reported to date, a comprehensive structural and biochemical study of the small subunit is necessary. However, so far, only the structure of the RRM domain from human U2AF35 has been solved (Kielkopf et al. 2001). Here we report the crystal structure of the whole fission yeast U2AF small subunit U2AF23 (in complex with the fragment of the large subunit U2AF59) (Kuhn and Käufer 2003), which exhibits the distinct binding activity and specificity for the 3′ splice site sequence. Together with functional analyses of directed mutant proteins, this structure shows a completely novel RNA recognition mechanism by the U2AF small subunit.
Results
The U2AF small subunit recognizes RNA by itself
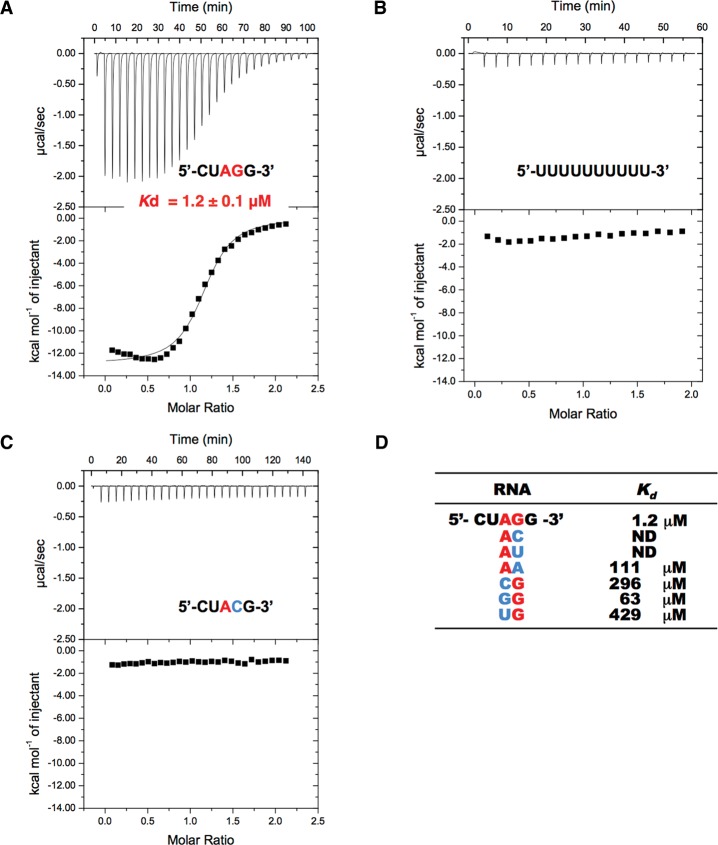
For the biochemical and structural study of the U2AF small subunit, we employed U2AF23 (composed of 216 amino acid residues), the small subunit of fission yeast Schizosaccharomyces pombe U2AF. U2AF23 is stabilized by binding to U2AF59, the S. pombe U2AF large subunit, so we purified U2AF23 as a complex (hereafter referred to as the “U2AF23 complex”) with a U2AF59 fragment spanning residues 93–161, which is longer than the above-mentioned 28-residue fragment (Fig. 1D). We carried out the isothermal titration calorimetry (ITC) experiments using the U2AF23 complex and different short RNAs. A 5-base ssRNA mimicking the 3′ splice site (5′-CUAGG-3′) and a 10-residue polyuridine tract were used as ligands, consistent with previous work with human U2AF35 (Wu et al. 1999). Upon addition of the U2AF23 complex to 5′-CUAGG-3′ RNA, heat release was observed, indicating simple one-to-one binding, with a Kd value of 1.2 μM (Table 1; Fig. 2A). In contrast, the polyuridine tract RNA did not cause any significant heat release (Table 1; Fig. 2B).
Table 1.
RNA-binding activities to the U2AF small subunit evaluated by ITC measurement
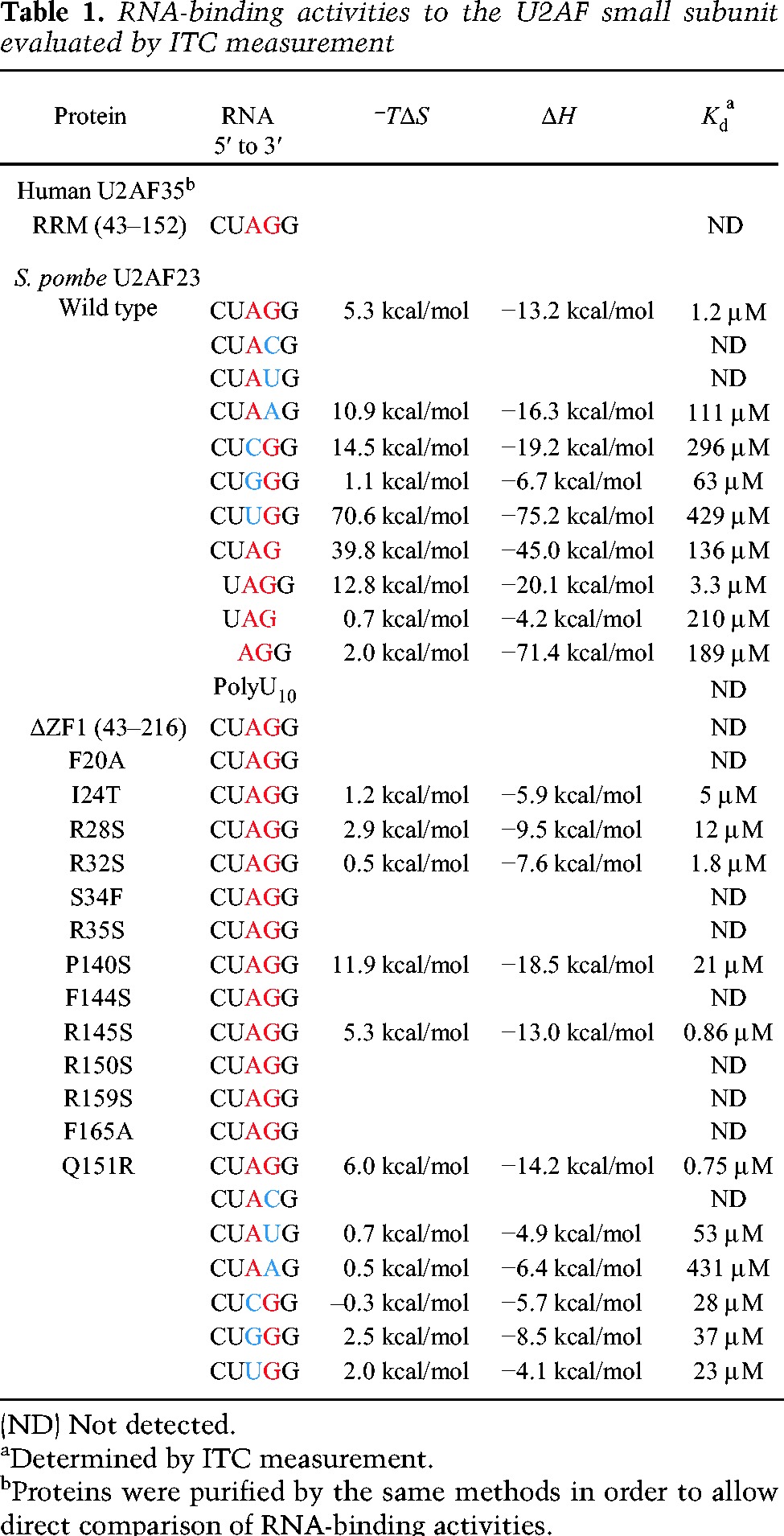
Figure 2.
RNA binding to the U2AF23 complex by ITC. Raw data of ITC measurements. The top panels show the heat release caused by injections of the U2AF complex into RNA CUAGG (A), UUUUUUUUUU (U10) (B), and CUACG (C). The bottom panels show the fitted binding parameters; the solid line in each bottom panel represents a calculated curve using the best fit parameters obtained by a nonlinear least squares fit. (D) RNA-binding activities calculated by ITC measurements using different RNA sequences. (ND) Not detectable.
We replaced the third A or fourth G nucleotides of the 5′-CUAGG-3′ sequence with other nucleotides and examined the effects on binding. In the RNA sequences with U or C nucleotides at the fourth position, no heat change could be detected; with adenosine at this position, the oligonucleotide showed a Kd value of 111 μM, within error, 100-fold higher than the Kd of the natural substrate sequence (Table 1; Fig. 2C,D). RNAs with C, G, and U at the third A position also exhibited much weaker binding affinities (Table 1; Fig. 2D). These results show unequivocally that the U2AF23 complex by itself recognizes the AG dinucleotide at the 3′ splice site in a sequence-specific manner.
Overall structure of U2AF23 in the complex
The crystal structure of the U2AF23 complex was solved and is shown in Figure 3A (the secondary structure elements are indicated in Fig. 1C). Regions spanning residues Lys15–Glu194 in U2AF23 and Arg105–Pro159 of U2AF59 showed clear electron density in the final map, but other parts of the complex were not found and were omitted from the model. The amino acid sequence shows that U2AF23 possesses a single RRM domain with a ZF domain on either side. Residues 15–43 and 143–170 correspond to the N-terminal and C-terminal ZFs, respectively (ZF1 and ZF2). The RRM domain has a canonical β1–α2–β2–β3–α3–β6 fold (Muto and Yokoyama 2012). The main chain structure of the U2AF23 RRM is closely superimposable on the model of the human U2AF35 RRM previously reported (Kielkopf et al. 2001), with a root mean square deviation (RMSD) value of 1.19 Å (Supplemental Fig. S1A). However, the two ZFs in U2AF23 lie against the β sheet of the RRM (Fig. 3A), which is the RNA-binding surface of canonical RRMs analyzed to date (Cléry et al. 2008). The tight binding of the three domains into a single structural unit shows that the U2AF23 RRM cannot bind to RNA directly in the usual manner. The C-terminal region of U2AF23 (spanning residues 171–194) after ZF2 adopts a long α-helical structure (α6 helix) parallel to the α2 helix in the RRM (Fig. 3A), and these two helices interact with each other through hydrophobic contacts.
Figure 3.
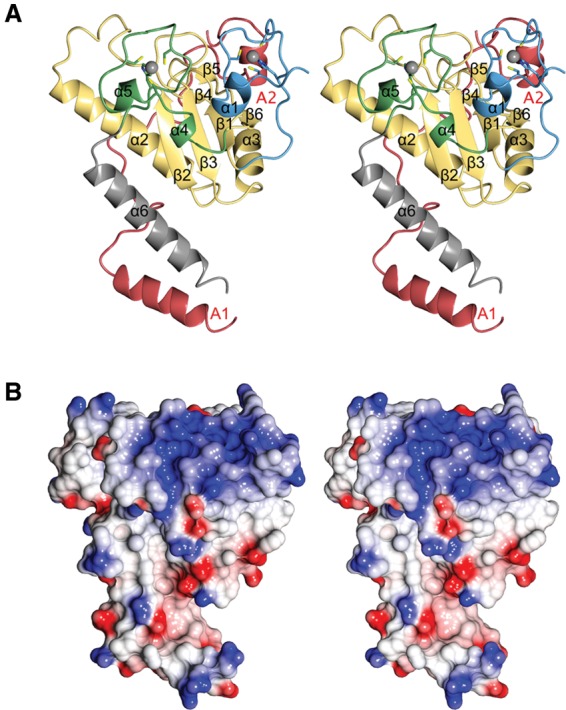
Crystal structure of the S. pombe U2AF small subunit complex with a short fragment of the U2AF large subunit. (A) Stereo ribbon representation of the overall structure of U2AF23 in the complex with the U2AF59 fragment. In U2AF23, the N-terminal ZF (ZF1, 15–43), RRM (44–141), C-terminal ZF (ZF2, 143–170), and U2AF59 fragment (105–159) are colored as in Figure 1A. Zinc ions are shown as gray spheres. Helices of U2AF23 are shown as ribbons and are numbered from the N terminus. The U2AF59 fragment forms two helices: A1 and A2. (B) Stereo view of the electrostatic surface of U2AF23 (red and blue indicate the negative and positive charge, respectively), shown from the same view as in A.
Interaction between U2AF23 and U2AF59
The U2AF23 RRM interacts with the region spanning residues 130–142 in the U2AF59 fragment through “reciprocal Trp recognition” mediated by Trp128 of U2AF23 and Trp135 of U2AF59 in the same manner as observed in the RRM domain of human U2AF35 (Supplemental Fig. S2A).
Furthermore, an important subunit contact not identified in the previous human U2AF35 RRM structure is found between the extended N-terminal region of the U2AF59 fragment (spanning residues 108–130), including the A1 helix (spanning residues 109–118) and the C-terminal α6 helix of U2AF23 (Fig. 1D; Supplemental Fig. S2B). Both of the helical regions were missing in the previous structural analysis of the U2AF35 RRM (Kielkopf et al. 2001). The newly discovered contact with the U2AF59 fragment lies over a wide area on the α2 and α6 helices of U2AF23. Hydrophobic amino acid residues of the N-terminal region of U2AF59 (Ile112, Leu116, Leu119, Val122, Ile125, and Trp128) form van der Waals contacts with the hydrophobic patch on the α6 helix of U2AF23, composed of Leu179, Leu181, Ala182, Tyr186, Ala188, and Ala191. The alkane chain of U2AF23 Lys185 is surrounded by the hydrophobic side chains of U2AF59 Leu119, Val122, and Pro124. U2AF59 Ile125 and Trp128 form a buried hydrophobic region with the α2 and α6 helices of U2AF23 (Supplemental Fig. S2B). These hydrophobic interactions are assisted by electrostatic interactions between U2AF23 Arg184 and U2AF59 Glu115 and other intersubunit hydrogen bonds (Supplemental Fig. S2B). Coexpression of the U2AF59 fragment (93–161) and the U2AF23 construct, including the α6 helix, gave higher expression and purification yields than U2AF complexes that could not form the complete set of interactions between the partner proteins (Supplemental Fig. S2C).
Two ZF domains are fixed on the RRM of U2AF23
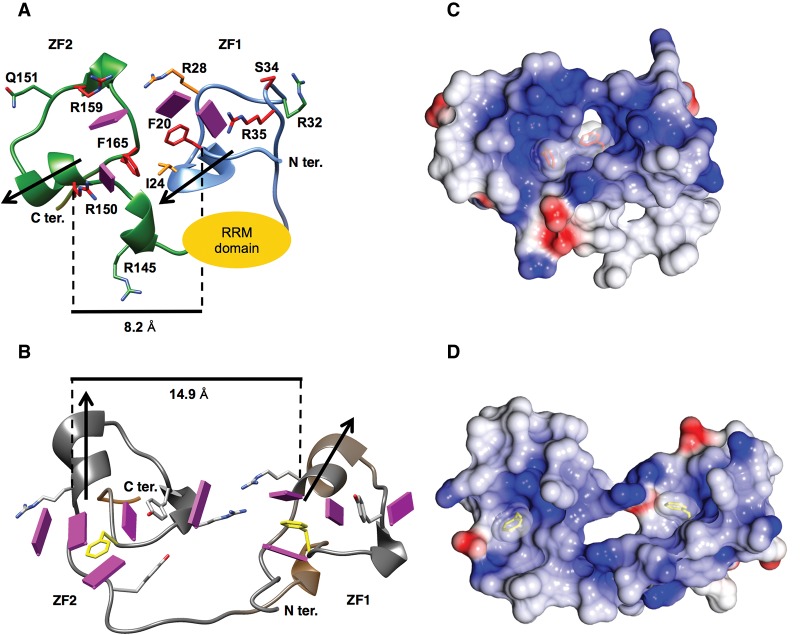
Two zinc ions were identified in this electron density map (Fig. 3A; Hall 2005; Mollet et al. 2006). One is coordinated by Cys18, Cys27, Cys33, and His37 of ZF1, and the other is coordinated by Cys149, Cys157, Cys163, and His167 of ZF2 (Figs. 1C, 3A). ZF1 obeys the standard pattern Cys-X8-Cys-X5-Cys-X3-His found in proteins such as TIS11d. ZF2 is different, with a pattern Cys-X7-Cys-X5-Cys-X3-His, which immediately follows Cys148. Although Cys148 is the first cysteine of the consensus motif, it does not coordinate the zinc atom and is replaced by Cys149. This is consistent with the previous mutational results, and we believe it is the only such case of a cysteine in such a ZF motif not in fact coordinating the metal (Webb and Wise 2004). The overall structures of both ZFs of U2AF23 resemble those of typical CCCH-type ZF family members, such as TIS11d ZF1, ZF2 (1RGO) (Hudson et al. 2004) or MBNL1 ZF1, ZF2 (3D2S) (Fig. 4; Supplemental Fig. S1B; Teplova and Patel 2008). However, the two ZFs of U2AF23 interact with each other on the β-sheet surface of the RRM domain by making an intimate mutual contact that has not been seen previously (Figs. 3A, 5).
Figure 4.
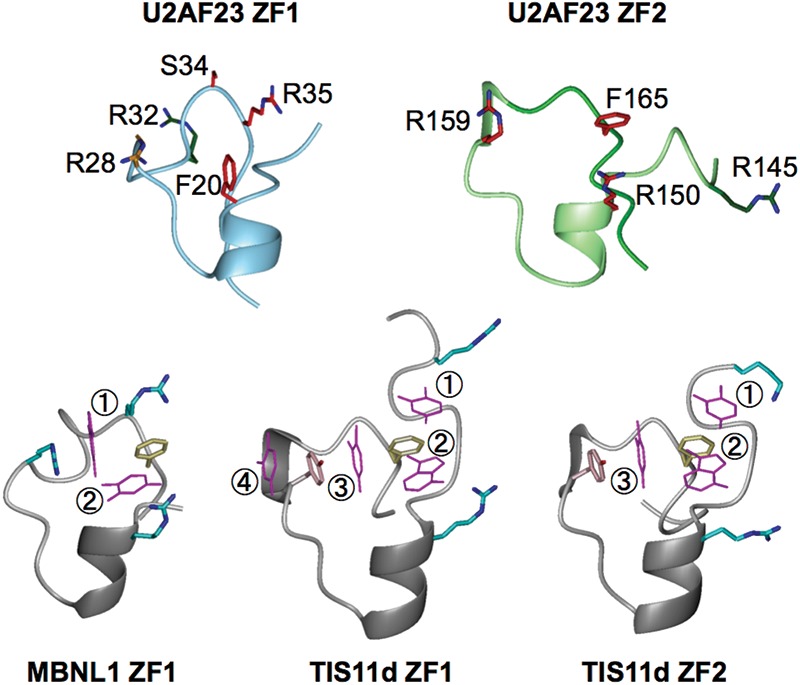
Structural comparison of CCCH-type ZFs. (Top panel) The structures of U2AF23 ZF1 (left) and ZF2 (right) shown as Cα traces decorated with side chains tested for involvement in RNA binding. The regions spanning residues 15–41 and 144–169 are demonstrated for U2AF23 ZF1 and ZF2, respectively. The side chains of Phe20, Arg28, Arg34, Ser34, and Arg35 of ZF1 and Arg145, Arg150, Arg159, and Phe165 of ZF2 are shown with the carbon atoms colored according to the results of the single-site mutagenesis experiments (Table 1); red indicates complete loss of binding activity, orange shows weakened binding, and green indicates no substantial effect on the RNA-binding activity. (Bottom panel) Structures of MBNL1 ZF1 (left), TISlld ZF1 (middle), and TISlld ZF2 (right). The well-conserved Phe residue (yellow) and the surrounding Arg, Lys (cyan), and Tyr (pink) residues create pockets suitable for binding RNA bases (shown in magenta and numbered from 5′ to 3′).
Figure 5.
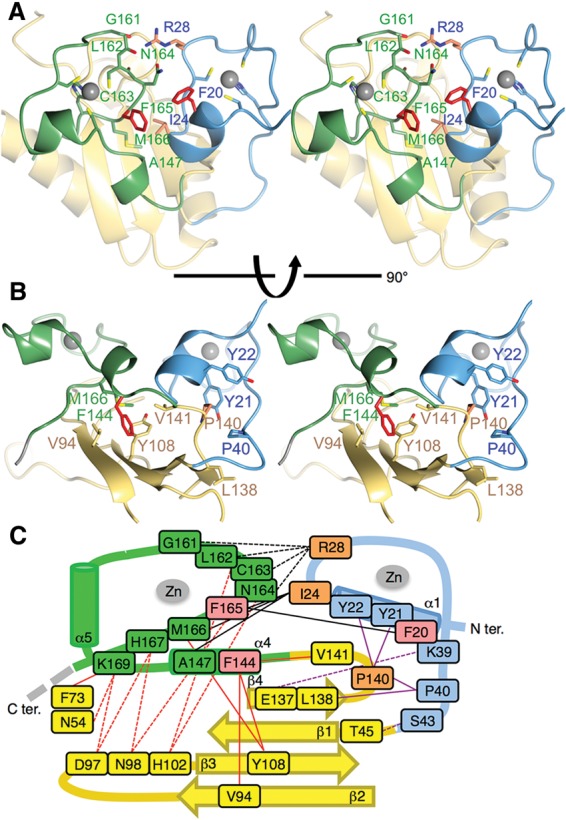
Interactions among ZF1, ZF2, and the RRM. (A) The interface between the ZF domains. The residues forming interactions between the two ZFs are shown as sticks and are colored as in Figure 4. (B) The interface between the ZFs and the RRM domains. Two Tyr residues of ZF1, Tyr21 and Tyr22, form the cage for P140, which is located just after the β6 strand of the RRM. Phe144 of ZF2 has hydrophobic contact with Val94, Tyr108, and Val141 of the RRM, and some hydrogen bonds stabilize the positions of the ZFs. (C) A schematic diagram showing the interactions between residues of the RRM and ZF domains in U2AF23. Hydrogen bonds are indicated by dotted lines, and hydrophobic contacts are indicated by solid lines. Interactions between domain partners are classified by different colors: ZF1 and the RRM in purple, ZF2 and the RRM in red, and ZF1 and ZF2 in black.
In detail, the side chain of Tyr21 on ZF1 extruded to the direction opposite to the RNA recognition surface of ZF1 and interacted with Pro140 on the RRM. Thus, Tyr21, Tyr22, and Pro40 of ZF1 form the cage for Leu137 and Pro140, which are located just after the β6 strand of the RRM (Fig. 5B,C). In addition, electrostatic interaction between Lys39 (ZF1) and Glu137 (RRM) and hydrogen bonds between Ser43 (ZF1) and Thr45 (RRM) stabilize the position of ZF1. On the other hand, Phe144 and Met166 of ZF2 have hydrophobic contact with Val94, Tyr108, and Val141 on the RRM and Ile24 of ZF1 (Fig. 5B,C). Furthermore, the two ZFs interact with each other through hydrophobic interactions among Phe20, Ile24 (ZF1), and Phe165 (ZF2) and through the hydrogen bonds from Arg28 of ZF1 to Gly161, Leu162, Cys163, and Asn164 of ZF2 (Fig. 5A,C). As a result, two ZFs of U2AF23 are closely fixed on the β-sheet surface of the RRM, and α1 (ZF1) and α5 (ZF2) helices are located on the line facing each other (Fig. 6A).
Figure 6.
Conformation of tandemly linked ZFs. Structures of ZF1 and ZF2 of U2AF23 (A) and of TIS11d (B). Arrows show the direction of α helices in which a third Zn-coordinating cysteine is located, and bars and lengths show the distances between these α helices. Pink plates are shown as RNA bases bound to ZFs; based on the model structure as an RNA–protein complex in A and on the solved structure of an RNA–protein complex in B. Electrostatic surfaces of U2AF23 (C) and that of TIS11d (D) shown from the same view as in A and B, respectively. Red and blue indicate the negative and positive charges, respectively. Residues forming a hydrophobic convex in U2AF23, Phe20, and Phe165 (orange) and nearly completely conserved aromatic amino acid residues (yellow) in TIS11d are shown.
Small-angle X-ray scattering (SAXS) measurements for the U2AF23 complex with or without RNA
SAXS studies were carried out with the U2AF23 complex in order to check whether the compact crystal structure is maintained in solution. The calculated radius of the particle (Rg) for the RNA-free form of the U2AF23 complex is 24.7 Å (Supplemental Fig. S3C). The calculated scattering curve from the crystal structure fits closely with the experimental result and Rg (Supplemental Fig. S3). Furthermore, the SAXS data indicate that the binding of 5′-CUAGG-3′ RNA to the U2AF23 complex does not bring about any major conformational change. These results indicate that the U2AF23 RRM domain remains covered by the two ZFs and is not surface-exposed. The previously unexpected result that the U2AF23 RRM domain does not bind directly to RNA matches our ITC experiments (Table 1).
Novel ZF1 of the U2AF23 complex
Canonical RNA-binding CCCH-type ZFs have an almost completely conserved central aromatic amino acid residue (Phe or Tyr) between the third and fourth Zn-coordinating residues in the conserved sequence Cx(F/Y)xH (Figs. 1B, 4), and this aromatic side chain plays an important role in stacking with RNA bases (Hudson et al. 2004; Teplova and Patel 2008). U2AF23 ZF1 lacks this aromatic residue, but ZF2 has it (Phe165) (Figs. 1B, 4). In TIS11d and MBNL1, this central side chain is surrounded by positively charged residues or tyrosine residues (Fig. 4), and nearby surface pockets can be identified that are suitable for binding the bases of substrate RNA. Bases projecting into these pockets interact with aromatic amino acid residues through π–π interaction (Hudson et al. 2004; Teplova and Patel 2008).
The mutant U2AF23 complex lacking ZF1 completely loses RNA-binding activity (Table 1), implying that the two ZFs function cooperatively and that ZF1 is necessary for RNA binding even though it lacks the generally conserved aromatic residue (Fig. 4). Another exposed aromatic residue, Phe20, is found in ZF1 at a position normally occupied by arginine in other CCCH-type ZFs (Fig. 4). Unique Phe20 of ZF1 plays an important role in the formation of a hydrophobic group with Ile24 of ZF1 and Phe165 of ZF2 at the center of a wide positively charged area (Fig. 6A,C). Therefore, unlike previously reported CCCH-type ZFs, U2AF23 ZFs combine to form a single RNA-binding surface, with the RRM domain holding them together rather than contacting the nucleic acid.
Putative RNA-binding sites on the U2AF23 complex
In order to confirm the role of residues within the positively charged surface, we performed several mutational experiments. First, we replaced the Phe20 and Phe165, which are in the center of the putative RNA-binding interface, with an alanine residue. The F20A and F165A mutations both completely abolished the RNA-binding activity of the U2AF23 complex (Table 1), confirming that these residues are functionally essential.
In the case of the canonical CCCH-type ZFs, positively charged surface residues create an RNA-binding site with the central Phe/Tyr residue. In the case of the U2AF23 complex, Arg28, Arg35, Arg150, and Arg159 are suitably positioned to form an RNA-binding pocket. Accordingly, we replaced each arginine with a serine residue and observed that the RNA-binding activity of the U2AF23 complex was completely abolished in the R35S, R150S, and R159S mutants (Table 1). In addition, the R28S mutant exhibited much weaker binding, with a Kd value of 12 μM, nearly 10-fold weaker than the wild-type U2AF23 complex (Table 1). These structural and functional data indicate that there are four pockets for base recognition, as shown in Figure 6A. The U2AF23 complex can bind the 4-base RNA 5′-UAGG-3′ with a Kd (3 μM) similar to that for the 5-base 5′-CUAGG-3′ RNA, but binding to shorter RNA molecules such as 5′-UAG-3′ or 5′-AGG-3′ is very weak (Table 1). Furthermore, binding of the U2AF23 complex to 5′-CUAG-3′ RNA is 100-fold weaker than that for 5′-UAGG-3′ RNA. These results indicate that the two ZFs of U2AF23 recognize a 4-base RNA in a sequence-specific manner, greatly preferring 5′-UAGG-3′.
These effects of single-site mutations on the RNA-binding activity of U2AF23 contrast strongly with tandemly linked CCCH-type ZFs (Michel et al. 2003; Hudson et al. 2004; Teplova and Patel 2008; Martinez-Lumbreras et al. 2013). In the case of the TIS11d protein, no single-site mutation on one ZF completely abolishes the RNA-binding activity of the protein overall because another ZF can still bind to the RNA molecule. The effects of mutating Phe20, Arg35, Arg150, Arg159, and Phe165 residues suggest strongly cooperative RNA recognition by ZF1 and ZF2. In contrast, putting a serine residue in place of Arg32 (ZF1) or Arg145 (ZF2), which are far from the hydrophobic convex, had no effect on the RNA-binding activity (Table 1).
In the U2AF23 complex, the ZFs are held together tightly on the β-sheet surface of the RRM (Fig. 5), and the interaction between the ZF and RRM seems to be crucial for the association of the two ZFs. In order to confirm the importance of the ZF–RRM interaction, we replaced Pro140 (of the RRM) and Phe144 (of ZF2) with a serine residue. These two residues mediate the interactions between ZF1 and the RRM domain and that between ZF2 and the RRM domain, respectively. For the P140S mutant, the RNA-binding affinity is decreased to 21 μM, and an even more dramatic loss of binding affinity is found for the F144S mutant (Fig. 5; Table 1). In addition, Ile24 is found at the interface between the ZFs, and RNA-binding activity decreases threefold for the I24T mutant (Table 1). Our structural analysis shows that these amino acid residues are not directly involved in the RNA binding but affect the affinity of binding indirectly. These results clearly demonstrate that cooperation between the two ZFs of U2AF23 is required for the RNA binding; the two ZFs create a unified positively charged area with hydrophobic knobs to form a single binding site for one consecutive RNA sequence.
Pathological mutation in the myelodysplastic syndromes
Recently, Yoshida et al. (2011) showed that patients with myelodysplastic syndromes often harbor specific mutations of U2AF35 at Ser34 and Gln157. Brooks et al. (2014) repeatedly identified certain somatic mutations in various types of cancer cells at Ile24, Ser34, and Gln157 in U2AF35. The three residues are conserved between humans and fission yeast (Gln157 in the human sequence corresponds to Gln151 in S. pombe) (Fig. 1C). The effect of the I24T mutation is described above, but we also examined mutations of the other two highly conserved amino acid residues in yeast U2AF23, which correspond to Ser34 and Gln157 in human U2AF35. The S34F mutant is strongly associated with pathogenesis and exhibits no RNA-binding activity (Table 1).
Not all mutants showed weakened RNA binding. The Q151R mutant exhibits stronger binding affinity for the CUAGG sequence, with a Kd value of 0.75 μM, despite being a pathogenic mutant (Table 1). We therefore examined the affinity of the Q151R mutant for several pentameric RNA sequences with different nucleotides at the third and fourth bases. Compared with the wild-type U2AF23 complex (with Kd values of 296, 63, and 429 μM for the CUCGG, CUGGG, and CUUGG sequences, respectively), the Q151R mutant showed rather strong binding affinities with a Kd value of ∼30 μM for these altered RNA sequences (Table 1). In addition, while the wild-type U2AF23 complex shows no significant binding to the RNA sequences with pyrimidine at the fourth position (guanine in the natural substrate), the Q151R mutant binds to the CUAUG sequence with a Kd value of 53 μM (Table 1). These results clearly show that the binding specificity of the U2AF23 complex is greatly weakened by the Q151R mutation, and the fact that this mutation is found to be associated with myelodysplastic syndromes can be attributed to the loss of splicing accuracy.
Discussion
Previously, several groups have shown the importance of the recognition of the AG dinucleotide at the 3′ splice site in the splicing reaction and that the U2AF small subunit strongly contributes to this binding. However, no structural understanding of this recognition has yet been reported, possibly in part because the human U2AF small subunit is too unstable for biological and biochemical measurements. In this study, we succeeded in preparing a stable form of the core part of the AG recognition module of the U2AF small subunit from fission yeast. The crystal structure described here shows a novel subunit contact between the C-terminal α6 helix of U2AF23 and the U2AF59 fragment, distinct from the “reciprocal Trp recognition” that is seen in the earlier model of the human U2AF35 RRM complex (Kielkopf et al. 2001). This additional subunit interaction stabilizes the whole yeast U2AF23 complex (Supplemental Fig. S2C), but the C-terminal α6 helix of U2AF23 is not highly conserved in higher vertebrates (Fig. 1C). Their U2AF small subunit has a C-terminal RS-rich sequence (RS domain) instead of the α6 helix (Fig. 1A). This RS domain of the eukaryotic U2AF small subunit plays an important role in the interaction with other proteins and/or RNA factors, and, in higher vertebrates, it seems probable that this domain strengthens binding between the U2AF large and small subunits with the assistance of other factors.
The present crystal structure shows novel module interactions that are crucial for specific RNA recognition by the U2AF small subunit. Two ZFs of U2AF form a single RNA-binding site, making intimate contact with each other, with one α helix from each ZF (the α1 and α5 helices) lying in parallel (Fig. 6A). In TIS11d, two ZFs are tandemly linked in the sequence but are more separated in space than the ZFs in the U2AF23 complex (Fig. 6A,B); these ZFs bind their target RNA molecules independently.
The yeast splicing factor Cwc2 also has a CCCH-type ZF and an RRM domain (McGrail et al. 2009). However, in the Cwc2 structure, the ZF is located on the opposite, α-helical side of the RRM, whose exposed β-sheet surface forms the RNA-binding site, as with other canonical RRMs. Schmitzová et al. (2012) showed that putative RNA-contacting sites map onto both the RRM and ZF domains. In the U2AF23 complex, the spatial relationship between the ZFs and RRM is completely novel, and the functional relationship between the structural elements are very different from previously described RNA-binding proteins.
The functional analysis of mutants together with the structural information identified three critical and one auxiliary Arg residues (Arg35, Arg150, and Arg159, and Arg28, respectively), which form 4-base recognition sites with the help of Phe20 and Phe165. An RNA-bound form of the U2AF23 complex with the 5′-UAGG-3′ RNA sequence could be modeled on the basis of our ITC results and the domain configuration of the MBNL1 protein (Fig. 7A). This model of the U2AF23 complex matches very well with the SAXS data as well as in the absence of RNA (Supplemental Fig. S3). It explains how three or four bases could be recognized by the U2AF23 complex on a unique RNA recognition surface created by interlocked ZFs on the RRM domain as well as why several somatic mutations in human U2AF35 at Ile24, Ser34, and Gln157 are commonly found in various types of cancer (Yoshida et al. 2011; Imielinski et al. 2012; Brooks et al. 2014; Shao et al. 2014). Our crystal structure shows that Ile24 is involved in the interaction between ZFs, which is essential for the cooperative formation of the RNA-binding site (Fig. 5A). The S34F mutant cannot bind to RNA (Table 1) because the bulky aromatic side chain at this position disturbs Arg35, leading to the loss of the RNA-binding activity. On the other hand, the Q151R mutant exhibits stronger binding ability for 5′-CUAGG-3′ RNA but with significant loss of sequence selectivity (Table 1). The structure of the RNA-free form of the U2AF23 complex shows a hydrogen bond between the side chains of Gln151 and Arg150. In the Q151R mutant, electrostatic repulsion between the two Arg residues disturbs the hydrogen-bonding network required for ligand selectivity.
Figure 7.
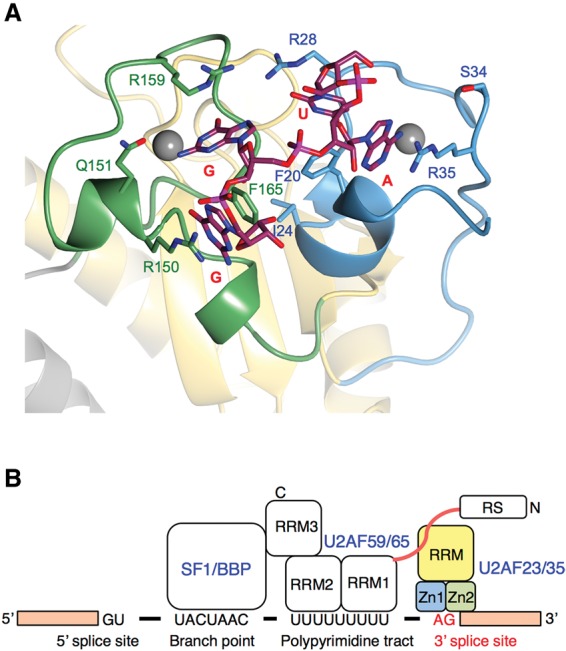
Novel 3′ splice site recognition by the U2AF small subunit. (A) Model of the RNA-bound U2AF23 complex based on the structural and mutational data. The demonstrated structure is the model of U2AF23 bound to a 4-base ssRNA mimicking the 3′ splice site 5′-UAGG-3′ on its ZFs. The mutation experiment along with the structural information could identify the three critical (Arg35, Arg150, and Arg159) and one auxiliary (Arg28) Arg residues that could form the base recognition site with the hydrophobic patch composed of Phe20 and Phe165. Based on the complex structure of the MBNL1 protein with the GC-containing RNA, the space between Arg159 and Phe165 and that between Phe165 and Arg150 (both belonging to ZF2) are assumed to be used for the successive guanine–guanine or guanine–cytidine sequences deduced from the putative hydrogen bonds. Instead, Phe20 of ZF1 contacts with Phe165 of ZF2, and we could find the space between Phe20 and Arg35 and that between Phe20 and Arg28. Based on the mutation experiment, Arg35 plays a more crucial role in the recognition of CUAGG RNA than Arg28. In addition, the space between Phe20 and Arg28 is considered to be in close proximity to the U2AF59 RRM1–RRM2 domains that bind to the PPT preceding the AG dinucleotide. Thus, we assumed that the space between Phe20 and Arg35 is for an adenine base and that the space between Phe20 and Arg28 is the auxiliary binding pocket for the nucleotides attached at the 5′ end of the adenine nucleotide. As described above, a hydrogen bond was formed between Arg28 of ZF1 and Asn164 of ZF2, and the side chain of Asn164 pointed to the binding pocket between Phe20 and Arg28. Thus, it is assumed that Asn164 is involved in the formation of the binding pocket, implying that the cooperation between ZF1 and ZF2 is necessary for the formation of the pocket. (B) Schematic representation of the 3′ splice site recognition mechanism by U2AF.
The present study reveals cooperative RNA binding by two firmly joined CCCH-type ZFs acting in concert to locate the AG dinucleotide at 3′ splice sites (Fig. 7B). This is the first evidence that the ZFs of the U2AF small subunit contribute directly to target site binding and recognition. Further structural study of the U2AF–RNA complex is needed to understand fully the regulation of the sequence-specific recognition of AG at the 3′ splice site by U2AF.
Materials and methods
Cloning, expression, and purification of the U2AF23–U2AF59 complex
The genes encoding U2AF23 and U2AF59 were amplified by PCR using fission yeast S. pombe genomic DNA. The product encoding U2AF59 residues 93–161 was digested with BamHI and NotI and then ligated into a suitably cut, modified pET28b vector in which an SD sequence, an initial ATG, a hexahistidine tag, and a tobacco etch virus (TEV) protease cleavage site had been cloned between the XbaI and BamHI restriction sites immediately upstream of the protein-coding region (pET28HisTEV). A U2AF23 gene was also cloned into modified pET28b using NdeI and NotI, and an NheI site was created by PCR between the stop codon and the NotI site. A coexpression plasmid was produced by ligating the hexahistidine-tagged U2AF59-coding fragment into NheI–NotI-digested pET28-U2AF23. This coexpression plasmid was transformed into the BL21(DE3)RIPL CodonPlus strain (Stratagene). The protein complex was expressed in LB medium overnight at 15°C after induction with 0.5 mM IPTG. Harvested cells were resuspended in Ni-NTA binding buffer (20 mM Tris at pH 8.0, 1 M NaCl, 500 mM urea, 25 mM imidazole, 10 mM β-mercaptoethanol) and lysed using EmulsiFlex homogenizer (Avestin). After centrifugation, the supernatant was loaded onto Ni-NTA agarose (Qiagen) equilibrated with the same buffer. Protein was eluted by a 25–500 mM linear gradient of imidazole. Peak fractions were incubated overnight with His-tagged TEV protease at room temperature while dialyzing against Ni-NTA low-salt buffer (20 mM Tris at pH 8.0, 150 mM NaCl, 25 mM imidazole, 10 mM β-mercaptoethanol). After complete cleavage, the sample was loaded on Ni-NTA agarose again to remove the His tag, His-tagged TEV protease, and minor protein contaminants. The complex was then dialyzed against crystallization buffer (20 mM Tris at pH 8.0, 100 mM NaCl, 1 mM DTT) and then concentrated to 20 mg/mL for crystallization using a Centriprep YM-10 concentrator (Millipore) with an exclusion size of 10 kDa. Mutagenesis was carried out by PCR. U2AF23 mutants were cloned and purified as complexes with the U2AF59 fragment by the same method. The genes encoding human U2AF35 and U2AF65 were amplified by PCR using cDNA (BC001177 and BC008740, respectively) in modified pET28b. The human U2AF complex was expressed and purified by the same methods.
Crystallization
Crystals were grown at 20°C in hanging drops that consisted of 1 µL of protein solution and 1 µL of reservoir solution, respectively. For the U2AF complex, 20 mg/mL protein in 20 mM Tris-HCl (pH 8.0) and 100 mM NaCl were crystallized with 25% PEG1000 and 0.1 M MES (pH 6.5) reservoir solution. Crystals were of space group C2221 with a = 91.6 Å, b = 112.7 Å, and c = 57.2 Å and contained one molecule in the asymmetric unit. Diffraction data were collected at −180°C using a crystal flash-frozen in crystallization buffer containing 30% (v/v) glycerol. Diffraction data from the native crystal were collected at 1.0 Å on the beamline Photon Factory BL1A using a Pilatus 1.2M-F detector. Diffraction data were integrated and scaled with HKL2000 and Scalepack (Otwinowski and Minor 1997). General handling of the scaled data was carried out with programs from the CCP4 suite (Collaborative Computational Project, Number 4 1994).
Structure determination and refinement
The structure was solved by molecular replacement using Phenix (Adams et al. 2010), with the human U2AF35 RRM domain (Protein Data Bank entry 1JMT) as the search model. The map was of good quality, allowing most of the model to be traced readily. Manual model building was performed using COOT (Emsley and Cowtan 2004), and refinement was carried out with Refmac (Murshudov et al. 1997) and Phenix (Adams et al. 2010). Solvent molecules were placed at positions where spherical electron density peaks were found above 1.3 σ in the |2Fo–Fc| map and above 3.0 σ in the |Fo–Fc| map and where stereochemically reasonable hydrogen bonds were allowed. The final model contained residues 15–194 of U2AF23 and residues 105–159 of the U2AF59 protein. Validation of the final model was carried out using Rampage (Lovell et al. 2003), which indicated that 98.7% of the residues are in the most favorable regions of the Ramachandran plot, with no residues in “disallowed” regions. A summary of the data collection and refinement statistics is given in Supplemental Table S1. Atomic coordinates and structure factors of the complex have been deposited in the Protein Data Bank under accession code 4YH8.
ITC experiments
All calorimetric titrations were carried out on VP-ITC and iTC200 calorimeters (MicroCal). RNAs were synthesized by Fasmac (Japan). Protein samples were dialyzed against the buffer containing 20 mM HEPES (pH 7.0) and 0.1 M NaCl. The sample cell was filled with a 50 μM solution of RNA, and the injection syringe was filled with 500 μM titrating U2AF complex wild type or mutants. For VP-ITC, each titration typically consisted of a preliminary 3-µL injection followed by 28 subsequent 10-µL injections every 210 sec. For iTC200, each titration typically consisted of a preliminary 0.4-µL injection followed by 19 subsequent 2-µL injections every 150 sec. All of the experiments were performed at 20°C. Data for the preliminary injection, which were affected by diffusion of the solution from and into the injection syringe during the initial equilibration period, were discarded. Binding isotherms were generated by plotting heats of reaction normalized by the moles of injected protein versus the ratio of the total injected one to total RNA per injection. The data were fitted using Origin software. All raw data using wild-type and mutant U2AF23 complexes are given in Supplemental Figure S4.
Modeling of the protein–RNA complex structure
Based on the results of ITC experiments of RNA and the amino acid substitution variant of U2AF, we arranged three nucleotides (A, G, G) in a row of A-G-G on the crystal structure of U2AF free-form and generated a variety of reasonable model structures. We further performed a total 1000 steps of energy minimization in a vacuum for these model structures with the Amber12 program: 500 of steepest descent (NCYC = 500) followed by 500 of conjugate gradient. The Amber ff99SB force field and 9999.0 Å cutoff, which means no cutoff, were applied.
SAXS experiments
SAXS measurements were performed at 4°C with a BioSAXS-1000 system (Rigaku) mounted on a MicroMax007HF X-ray generator (Rigaku). The Pilatus 100 k detector, at a sample-to-detector distance of 484 mm, was used to measure scattering intensities. Samples dissolved in 20 mM HEPES (pH 7.0) containing 0.5 M NaCl were used for SAXS measurement. The samples containing 1.5 equimolar 5-base ssRNA were used for analysis of the RNA-bound form. A circular averaging of the scattering intensities was calculated by using SAXSLab (Rigaku) to obtain one-dimensional scattering data I(q) as a function of q (q = 4πsinθ/λ, where 2θ is the scattering angle, and the X-ray wavelength λ = 1.5418 Å). To correct for interparticle interference, I(q) data were collected at three different protein concentrations (3.7 mg/mL, 2.4 mg/mL, and 1.1 mg/mL). Exposure time was 8 h for 3.7 mg/mL and 2 h for 2.4 mg/mL and 1.1 mg/mL. However, the intensity profile did not indicate a concentration effect; therefore, the correction for the interparticle interference was not applied. All SAXS data were analyzed with the software applications embedded in the ATSAS package (http://www.embl-hamburg.de/biosaxs/software.html). The radius of gyration Rg and forward scattering intensity I(0) were estimated from the Guinier plot of I(q) in a smaller angle region of qRg < 1.3 (Glatter 1982). The molecular mass of the U2AF23 complex was estimated by comparing its I(0) with those of ovalbumin and lysozyme. Models of disordered regions in the crystal structure were built by BilboMD (Pelikan et al. 2009). The I(q) of the crystal structure or model structures was calculated by using FoXS (Schneidman-Duhovny et al. 2010, 2013). χ2 values were calculated against an experimental I(q) at 3.7mg/mL on a q range of 0.018–0.251 (Å−1).
Supplementary Material
Acknowledgments
We thank the staff at the Photon Factory BL1A beamlines for assistance with data collection. We also thank Professor Jeremy Tame for critical reading of the manuscript. We acknowledge the technical expertise of the Center for Integrated Research in Science at Shimane University. We also thank Yuko Fukuma for technical assistance. This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas “RNA regulation” (no. 20112006) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and also by the SUIGAN Project, Shimane University.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.267104.115.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, Pedamallu CS, Sivachenko A, Rosenberg M, Chmielecki J, et al. 2014. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS One 9: e87361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cléry A, Blatter M, Allain FH. 2008. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 18: 290–298. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Glatter O. 1982. Data treatment. In Small-angle X-ray scattering (ed. Glatter O, Kratky O), pp. 119–165. Academic Press, New York. [Google Scholar]
- Guth S, Tange TO, Kellenberger E, Valcarcel J. 2001. Dual function for U2AF35 in AG-dependent pre-mRNA splicing. Mol Cell Biol 21: 7673–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TM. 2005. Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol 15: 367–373. [DOI] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. 2004. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11: 257–264. [DOI] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. 2012. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150: 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 12: 5–14. [DOI] [PubMed] [Google Scholar]
- Kielkopf CL, Rodionova NA, Green MR, Burley SK. 2001. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 106: 595–605. [DOI] [PubMed] [Google Scholar]
- Kuhn AN, Käufer NF. 2003. Pre-mRNA splicing in Schizosaccharomyces pombe: regulatory role of a kinase conserved from fission yeast to mammals. Curr Genet 42: 241–251. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB III, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. 2003. Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins 50: 437–450. [DOI] [PubMed] [Google Scholar]
- Martinez-Lumbreras S, Santiveri CM, Mirassou Y, Zorrilla S, Perez-Canadillas JM. 2013. Two singular types of CCCH tandem zinc finger in Nab2p contribute to polyadenosine RNA recognition. Structure 21: 1800–1811. [DOI] [PubMed] [Google Scholar]
- McGrail JC, Krause A, O'Keefe RT. 2009. The RNA-binding protein Cwc2 interacts directly with the U6 snRNA to link the nineteen complex to the spliceosome during pre-mRNA splicing. Nucleic Acids Res 37: 4205–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martinez C, Valcárcel J. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841. [DOI] [PubMed] [Google Scholar]
- Michel SL, Guerrerio AL, Berg JM. 2003. Selective RNA binding by a single CCCH zinc-binding domain from Nup475 (Tristetraprolin). Biochemistry 42: 4626–4630. [DOI] [PubMed] [Google Scholar]
- Mollet I, Barbosa-Morais NL, Andrade J, Carmo-Fonseca M. 2006. Diversity of human U2AF splicing factors. FEBS J 273: 4807–4816. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255. [DOI] [PubMed] [Google Scholar]
- Muto Y, Yokoyama S. 2012. Structural insight into RNA recognition motifs: versatile molecular Lego building blocks for biological systems. Wiley Interdiscip Rev RNA 3: 229–246. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, Hammel M. 2009. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys 28: 174–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Breger KS, Kanaar R, Adams MD, Rio DC. 1998. RNA binding activity of heterodimeric splicing factor U2AF: at least one RS domain is required for high-affinity binding. Mol Cell Biol 18: 4004–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitzová J, Rasche N, Dybkov O, Kramer K, Fabrizio P, Urlaub H, Lührmann R, Pena V. 2012. Crystal structure of Cwc2 reveals a novel architecture of a multipartite RNA-binding protein. EMBO J 31: 2222–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Sali A. 2010. FoXS: a Web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res 38: W540–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. 2013. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys J 105: 962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Yang B, Wu T, Huang J, Tang P, Zhou Y, Zhou J, Qiu J, Jiang L, Li H, et al. 2014. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat Struct Mol Biol 21: 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. 2006. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell 23: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares LM, Zanier K, Mackereth C, Sattler M, Valcárcel J. 2006. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science 312: 1961–1965. [DOI] [PubMed] [Google Scholar]
- Teplova M, Patel DJ. 2008. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol 15: 1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Webb CJ, Wise JA. 2004. The splicing factor U2AF small subunit is functionally conserved between fission yeast and humans. Mol Cell Biol 24: 4229–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. 2011. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478: 64–69. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Green MR. 1989. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci 86: 9243–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355: 609–614. [DOI] [PubMed] [Google Scholar]
- Zuo P, Maniatis T. 1996. The splicing factor U2AF35 mediates critical protein–protein interactions in constitutive and enhancer-dependent splicing. Genes Dev 10: 1356–1368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.