In Bacillus subtilis, condensin is loaded at centromeric parS sites, where it encircles DNA and individualizes newly replicated origins. Wang et al. show that condensin recruitment to origin-proximal parS sites is required for the juxtaposition of the two chromosome arms.
Keywords: lengthwise condensation, Hi-C, DNA segregation, SMC, mitosis, ParB, parS
Abstract
SMC condensin complexes play a central role in compacting and resolving replicated chromosomes in virtually all organisms, yet how they accomplish this remains elusive. In Bacillus subtilis, condensin is loaded at centromeric parS sites, where it encircles DNA and individualizes newly replicated origins. Using chromosome conformation capture and cytological assays, we show that condensin recruitment to origin-proximal parS sites is required for the juxtaposition of the two chromosome arms. Recruitment to ectopic parS sites promotes alignment of large tracks of DNA flanking these sites. Importantly, insertion of parS sites on opposing arms indicates that these “zip-up” interactions only occur between adjacent DNA segments. Collectively, our data suggest that condensin resolves replicated origins by promoting the juxtaposition of DNA flanking parS sites, drawing sister origins in on themselves and away from each other. These results are consistent with a model in which condensin encircles the DNA flanking its loading site and then slides down, tethering the two arms together. Lengthwise condensation via loop extrusion could provide a generalizable mechanism by which condensin complexes act dynamically to individualize origins in B. subtilis and, when loaded along eukaryotic chromosomes, resolve them during mitosis.
Compaction of replicated chromosomes into morphologically and spatially distinct sister chromatids is essential for faithful DNA segregation. In virtually all organisms, SMC condensin complexes play a central role in this process. These highly conserved ring-shaped complexes are composed of two SMC subunits, a kleisin (“closure”) family member, and accessory factors (Cuylen and Haering 2011; Hirano 2012). Since their discovery over two decades ago (Hirano and Mitchison 1994; Saka et al. 1994; Strunnikov et al. 1995), condensins have been found to constrain DNA (Kimura and Hirano 1997; Stray et al. 2005), bridge DNA segments (Petrushenko et al. 2010), and topologically embrace DNA helices (Cuylen et al. 2011; Wilhelm et al. 2015), yet the mechanism by which they compact and resolve replicated chromosomes has remained elusive.
In eukaryotes, condensins act at the earliest stages of mitosis to compact and resolve interphase chromosomes into rod-shaped structures that assemble at the metaphase plate (Hirano 2012). Recent studies underscore the structural and functional similarities between the Bacillus subtilis condensin complex and its eukaryotic counterparts. B. subtilis condensin is composed of SMC, ScpA (kleisin), and the accessory protein ScpB (Britton et al. 1998; Mascarenhas et al. 2002; Soppa et al. 2002). An SMC homodimer and ScpA form a tripartite ring that is similar in size, architecture, and intersubunit contacts to eukaryotic condensins (Hirano et al. 2001; Burmann et al. 2013; Kamada et al. 2013). In bacteria, chromosome segregation occurs concomitant with DNA replication, and B. subtilis condensin plays a central role in the earliest step in this process: the compaction and resolution (or individualization) of newly replicated sister origins (Gruber et al. 2014; Wang et al. 2014b). Inactivation of SMC during growth in rich medium leads to the accumulation of unresolved origins and a lethal defect in chromosome segregation. In slow growth conditions, cells lacking condensin subunits are viable but inefficiently individualize their origins and produce a high frequency of anucleated cells (Britton et al. 1998; Gruber et al. 2014; Wang et al. 2014b). These defects are significantly enhanced by mutations in the parABS partitioning system that actively segregates replicated origins toward opposite cell poles (Lee and Grossman 2006; Wang et al. 2014a,b).
In addition to promoting origin segregation, the ParB partitioning protein bound to centromeric parS sites also functions to recruit condensin to its site of action (Gruber and Errington 2009; Sullivan et al. 2009). Cells lacking ParB or its eight origin-proximal parS sites (but not the ParA ATPase) have a significant reduction in origin-localized condensin. Furthermore, recent studies indicate that B. subtilis condensin entraps chromosomal DNA in vivo, and this entrapment is strongly reduced in the absence of ParB, suggesting that most condensin is loaded onto the chromosome at parS sites (Wilhelm et al. 2015). Despite ParB's role in recruiting condensin to the origin, cells lacking ParB have a modest (fivefold to 80-fold) increase in anucleated cells (Ireton et al. 1994; Lee and Grossman 2006), suggesting that the limited number of condensin complexes loaded in the absence of ParB (Wilhelm et al. 2015) is sufficient to resolve replicated origins. However, when combined with hypomorphic alleles of SMC, ParB mutants have severe defects in origin resolution and chromosome segregation (Gruber and Errington 2009; Wang et al. 2014b). Thus, the current thinking is that ParB loads condensin at parS sites, where it individualizes newly replicated origins (Gruber and Errington 2009; Wang et al. 2014b). Once resolved, they are actively segregated by ParA acting on ParB/parS. As is the case in eukaryotes, the mechanism by which B. subtilis condensin resolves sister origins is unknown.
Here, we investigated the contribution of ParB, the eight origin-proximal parS sites, and condensin to the organization of the chromosome using chromosome conformation capture (3C) combined with deep sequencing (Hi-C) (Lieberman-Aiden et al. 2009). We show that interactions between the two chromosome arms and between the origin region and sites along the left and right arms require all three components. A single parS site in the origin region is sufficient for interarm interaction and specifies the inflection point for the juxtaposition of the two arms. Strikingly, insertion of an ectopic parS site at an origin-distal position does not restore interarm interactions but instead promotes condensin-dependent alignment of large tracks of DNA on either side of parS. Analysis of chromosome conformation in cells with two parS sites on opposing arms revealed that these “zip-up” interactions occur along adjacent DNA segments and not between DNA on opposite arms. Finally, Hi-C analysis of cells with 1-N content that are blocked for replication initiation indicates that the closely spaced origin-proximal parS sites similarly promote alignment of flanking DNA, generating long-range interactions that emanate from the origin region. Altogether, these data support a generalizable model in which condensin resolves replicated origins by promoting the juxtaposition of flanking DNA, drawing sister origins in on themselves and away from each other.
Results
Global changes in chromosome conformation in cells lacking ParAB
To probe the organization of the Bacillus subtilis chromosome at high resolution, we performed Hi-C on wild-type cells during exponential growth in rich medium (see the Materials and Methods). For all of the experiments presented, we used the endonuclease HindIII that generates cleavage products with an average fragment size of 1.7 kb. To analyze the resulting data, the chromosome was divided into 10-kb bins, and the probability of contact between each pair of restriction fragments was assigned to a corresponding bin. To more clearly visualize interactions in the origin region, unlike previously reported Hi-C contact probability matrices for bacterial chromosomes (Umbarger et al. 2011; Le et al. 2013; Marbouty et al. 2014), the genome was oriented along the X- and Y-axes with the origin at the center and the left and right arms on either side (Fig. 1A; Supplemental Fig. S1). As observed using metagenomic 3C (meta-3C) at lower resolution (Marbouty et al. 2014), wild-type B. subtilis cells displayed extensive short-range interactions along both chromosome arms (the primary diagonal in the contact matrix) and weaker but robust interactions between the two arms (the secondary diagonal) (Fig. 1A; Supplemental Fig. S1). For most of the replication–segregation cycle, the B. subtilis chromosome adopts an “ori-ter” configuration in which the left and right arms lie side by side (Wang et al. 2014a). The Hi-C performed here was on asynchronously growing cells and was likely dominated by chromosomes in this configuration, accounting for the extensive interarm interactions observed.
Figure 1.
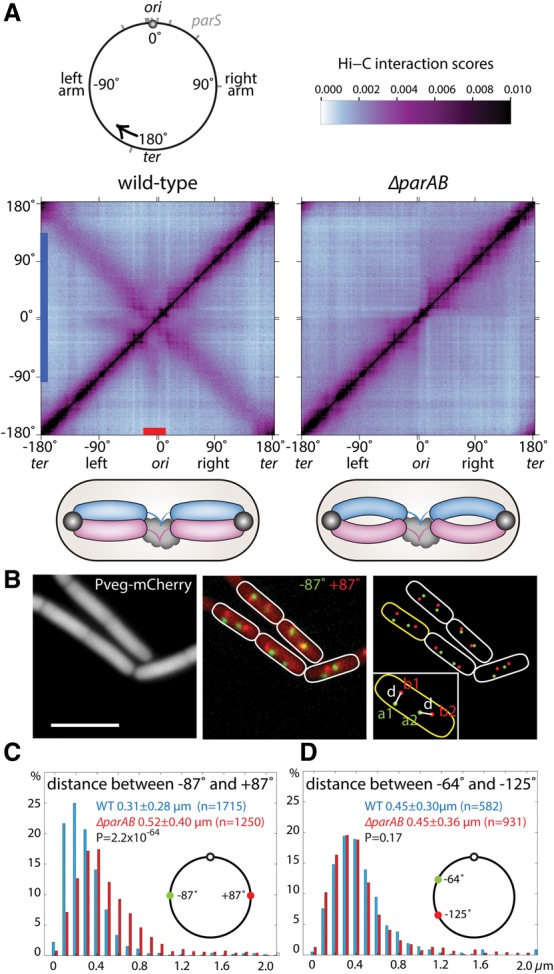
Long-range chromosome interactions require the parAB locus. (A) Normalized HindIII Hi-C interaction matrices displaying contact frequencies for pairs of 10-kb bins across the 4.0-Mb genome for wild-type (PY79) and ΔparAB (BDR1873) cells grown at 37°C in rich medium. Axes indicate the genome position of each bin in degrees. To more clearly visualize interactions in the origin region, the genome was oriented along the axes with the origin (ori) at the center and the left and right arms on either side (see Supplemental Fig. S1). An ∼300-kb region spanning the origin (red bar on the X-axis) and a broad region on the left and right arms (blue bar on the Y-axis) that have long-range interactions in wild-type are highlighted (see Supplemental Fig. S3A). The top left panel shows the circular B. subtilis chromosome with positions of the 10 parS sites (gray bars). The black arrow indicates the start and direction of the axes on the Hi-C contact maps. The top right panel has the color scale bar for all Hi-C contact maps presented in this study. Below the contact maps are schematics showing the ori-ter-ori configuration (Wang et al. 2014a) in B. subtilis cells and the presence or absence of interarm interactions in wild type and ΔparAB. Origins (black balls), chromosomal arms (blue and purple bars), and unreplicated DNA (black clouds) are shown. Long-range interactions emanating from the origin region are not shown. (B) Representative example of image analysis. Cytoplasmic mCherry was used to determine the outlines of the cells. Chromosomal loci at +87° and −87° were visualized using tetO/TetR-CFP and lacO/LacI-mYpet (BWX1912). Bar, 4 µm. (C) The distance between loci on opposing arms increases in cells lacking parAB (BWX3168) compared with wild type (BWX1912). (D) The distance between loci on the same arm remains unchanged in cells lacking parAB (BWX3446) compared with wild type (BWX3248). The X-axis is the interfocal distance in micrometers. The Y-axis is the percentage of cells that fall in each 0.1-µm bin. P-values were calculated using two-sample Kolmogorov-Smirnov test in Matlab. Details of analysis are in the Supplemental Material. Analyses of cells growing in minimal medium or cells lacking parA are in Supplemental Figure S3.
Similar to what has been reported in Caulobacter crescentus (Le et al. 2013), chromosome interaction domains (CIDs) of 60–340 kb were present along each arm (Fig. 1A; Supplemental Fig. S2), and many of these domains were bounded by highly transcribed genes. Although the boundaries were significantly less pronounced than those observed in C. crescentus, they were present in more than four independent Hi-C experiments. Furthermore, several of these domains became significantly more pronounced in specific mutant backgrounds that are presented below.
A striking feature in the contact matrix of wild-type cells that was not observed in C. crescentus was the presence of long-range interactions between an ∼300-kb region of the origin and loci spanning up to 1.5 Mb on both the left and right arms (Fig. 1A; Supplemental Fig. S3A). This region spans most of the origin-proximal parS sites that are bound by ParB, forming large nucleoprotein complexes that are acted on by ParA (Murray et al. 2006; Breier and Grossman 2007; Wang et al. 2014a). To investigate whether these interactions require the ParABS partitioning system, we performed Hi-C on a strain lacking the parAB locus. Strikingly, the long-range interactions emanating from the origin region were completely absent in the mutant, while the short-range interactions along the chromosome arms were largely unchanged (Fig. 1A; Supplemental Fig. S2A,B). However, the most dramatic change in the conformation of the chromosome in cells lacking ParAB was the loss of interaction between the two chromosome arms (Fig. 1A). Instead, each arm formed independent interaction domains separated by the highly transcribed genes in the origin region. Interarm interactions were similarly lost in ΔparAB cells during slow growth in minimal medium (Supplemental Fig. S3E) and during the developmental process of spore formation (Supplemental Fig. S3G).
We note that wild-type chromosomes adopt distinct conformations in the three growth conditions tested with common and unique short- and long-range interactions. These will be the subject of a separate study. The principal focus of the work presented here is the interactions between the two chromosome arms and between the origin region and distal sites (collectively referred to as long-range interactions).
The distance between loci on opposing arms increases in the absence of ParAB
To investigate whether the loss of interarm interaction in the ΔparAB mutant reflects the disposition of the chromosome in single cells, we inserted tetO and lacO arrays on opposing arms of the chromosome (at ±87°) and used fluorescently tagged repressor proteins to monitor their localizations (Wang et al. 2014a). We collected snapshot images of wild-type and mutant cells grown in rich and minimal media and determined interfocal distances using automated image analysis (Fig. 1B; see the Supplemental Material; Sliusarenko et al. 2011). Consistent with the loss of interarm interaction observed by Hi-C, the distances between loci on opposing arms increased significantly in the ΔparAB mutant compared with wild type under both growth conditions (Fig. 1C; Supplemental Fig. S3F). Furthermore, interfocal distances between fluorescently tagged loci on the same chromosome arm (at −64° and −125°) were similar in the two strains (Fig. 1D), consistent with the Hi-C data, indicating that short-range interactions along the chromosome arms were largely unchanged in the absence of parAB (Supplemental Fig. S2A,B).
ParB/parS nucleoprotein complexes are required for long-range interactions
In B. subtilis, the ParABS system promotes the segregation of replicated origins (Wang et al. 2014a), while ParB bound to parS additionally functions to load the condensin complex in the origin region (Gruber and Errington 2009; Sullivan et al. 2009; Wilhelm et al. 2015). To determine which of these activities is required for the long-range interactions observed by Hi-C, we analyzed strains lacking parA, parB, or nine parS sites (eight origin-proximal parS sites at −1°, −4°, −5°, −6°, −27°, +4°, +17°, and +42° and the origin-distal site at +91°, referred to as parSΔ9) (Lin and Grossman 1998; Breier and Grossman 2007). The long-range interactions that required ParAB were completely absent in cells lacking ParB and the nine parS sites but remained largely unchanged in the absence of ParA (Fig. 2A; Supplemental Figs. S3A,B, S4). Consistent with the idea that ParA is not required for interarm interactions, ΔparA mutant cells had a very modest increase in the distance between +87° and −87° loci compared with cells lacking the parAB locus (Fig. 1C; Supplemental Fig. S3C,D,F).
Figure 2.
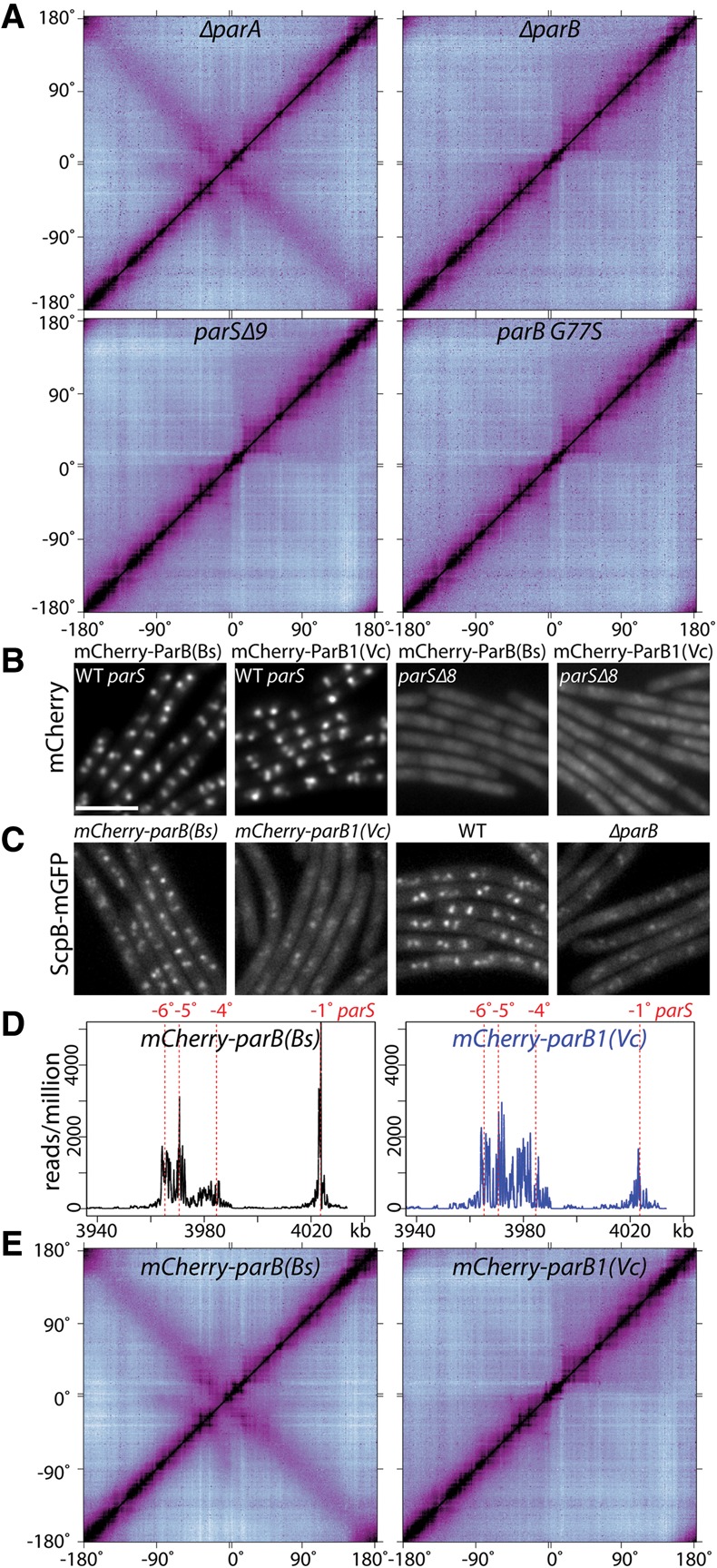
Long-range chromosome interactions require condensin recruitment to ParB/parS nucleoprotein complexes. (A) Normalized Hi-C contact matrices for cells lacking parA (BWX2876), parB (BDR2292), nine parS sites (parSΔ9; BWX3196), or a strain expressing ParB G77S (BDR2707) that binds parS but cannot form higher-order complexes. (B–E) ParB1 from Vibrio cholerae [ParB1(Vc)] forms nucleoprotein complexes at parS sites in B. subtilis but does not recruit condensin or support long-range chromosomal interactions. (B) Localization of mCherry-ParB(Bs) and mCherry-ParB1(Vc) in cells with the full complement of parS sites (BWX3208 and BWX3209) or cells lacking eight origin-proximal parS sites (parSΔ8; BWX2519 and BWX2520). (C) Localization of ScpB-mGFP in cells expressing mCherry-ParB(Bs) (BWX2514), mCherry-ParB1(Vc) (BWX2515), wild-type ParB (BWX2030), or no ParB (ΔparB) (BWX2049). Fluorescence intensity was scaled to the same level for each channel. Bar, 4 µm. (D) Enrichment of mCherry-ParB(Bs) (BWX3208) and mCherry-ParB1(Vc) (BWX3209) as assayed by chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) at a region of the B. subtilis chromosome (from 3940 to 4033 kb) encompassing four parS sites (at −1°, −4°, −5° and −6°). The number of reads per million reads was plotted on the Y-axis. mCherry-ParB(Bs) and mCherry-ParB1(Vc) spread to similar extents. ChIP-seq profiles for all of the individual parS sites and the entire genome are in Supplemental Figure S5. (E) Normalized Hi-C contact matrices of ΔparB mutants expressing mCherry-ParB(Bs) (BWX3208) or mCherry-ParB1(Vc) (BWX3209).
ParB proteins form large nucleoprotein complexes at parS sites that span 10–20 kb of flanking DNA (Murray et al. 2006; Breier and Grossman 2007) mediated by nearest neighbor and bridging interactions (Graham et al. 2014; Taylor et al. 2015). To determine whether these nucleoprotein complexes are required for long-range interactions, we performed Hi-C on cells harboring a ParB point mutant (G77S) that binds parS sites with affinity similar to wild type but does not spread to neighboring sites (Breier and Grossman 2007; Graham et al. 2014). The interaction map for this mutant resembled those derived from the ΔparB and parSΔ9 strains (Fig. 2A; Supplemental Fig. S4). Collectively, these results indicate that ParB nucleoprotein complexes assembled at parS sites are required for interarm interactions along the length of the chromosome arms and long-range interactions between the origin region and distal sites.
ParB-mediated recruitment of condensin is required for long-range interactions
ParB/parS nucleoprotein complexes are required to recruit condensin to the origin of replication (Gruber and Errington 2009; Sullivan et al. 2009). To determine whether the ParB/parS nucleoprotein complexes or condensin itself was required for the long-range interactions, we used a B. subtilis strain harboring Vibrio cholerae ParB1 [referred to as ParB1(Vc)] that forms nucleoprotein complexes at parS sites but is unable to recruit condensin. ParB1(Vc) recognizes the identical parS site in vivo (Livny et al. 2007), and single-molecule experiments in vitro indicate that ParB1(Vc) mediates nearest neighbor and bridging interactions similar to B. subtilis ParB (Graham et al. 2014). Figure 2B shows that B. subtilis cells harboring an mCherry-ParB1(Vc) fusion in a strain lacking the endogenous parB gene generated fluorescent foci that depend on the origin-proximal parS sites. These foci are similar to those formed by an mCherry fusion to the B. subtilis ParB protein [called mCherry-ParB(Bs)] (Fig. 2B). Furthermore, ChIP-seq (chromatin immunoprecipitation [ChIP] combined with deep sequencing) experiments using anti-mCherry antibodies indicate that both fusion proteins are similarly enriched at origin-proximal parS sites and along adjacent DNA segments (Fig. 2D; Supplemental Fig. S5). Thus, ParB1(Vc) and B. subtilis ParB likely generate similar nucleoprotein complexes in vivo. Importantly, cells harboring mCherry-ParB1(Bs) but not mCherry-ParB1(Vc) had discrete condensin complexes, as assayed by a GFP fusion to the ScpB subunit (ScpB-mGFP) (Fig. 2C). Thus, a ParB/parS nucleoprotein complex is necessary but not sufficient to recruit condensin, and ParB1(Vc) can serve as a separation-of-function mutant. We performed Hi-C on B. subtilis cells expressing mCherry-ParB1(Vc) and separately on those expressing mCherry-ParB(Bs). Consistent with the idea that condensin recruitment is required for long-range interactions, the contact map from cells expressing mCherry-ParB1(Vc) was similar to the parB null (Fig. 2A,E; Supplemental Fig. S4).
Condensin is required for long-range interactions
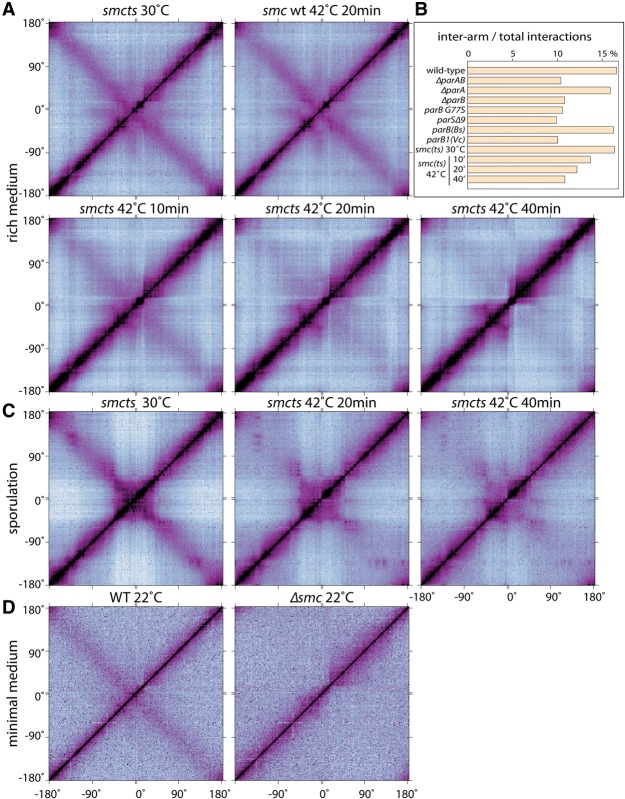
The condensin complex is conditionally essential in B. subtilis: It is required for fast growth in rich medium but is dispensable under slow growth conditions. In its absence, origin resolution and chromosome segregation are impaired, resulting in heterogeneous chromosome morphologies (Wang et al. 2014b). Accordingly, to assess whether condensin is required for long-range interactions, we used a temperature-sensitive smc allele (smcts) to rapidly inactivate the protein (Wang et al. 2014b). Cells were grown under permissive conditions (30°C) in defined rich medium and then shifted to the restrictive temperature (42°C). Samples were taken before and 10, 20, and 40 min after the shift to the nonpermissive temperature and analyzed by Hi-C. Under permissive conditions, the contact map was similar to the one generated from wild-type cells grown at 37°C or 42°C (Figs. 1A, 3A; Supplemental Fig. S4B). In contrast, the interactions between the two arms and between the origin and distal sites were progressively lost after shifting the mutant to the nonpermissive temperature (Fig. 3A,B; Supplemental Fig. S6). One caveat of this experiment is that, shortly after SMC is inactivated, newly replicated origins fail to individualize, and the replicated chromosomes remain unsegregated (Gruber et al. 2014; Wang et al. 2014b). Although the long-range interactions observed by Hi-C were significantly reduced after only 10 min at the restrictive temperature, it was formally possible that the loss of interaction was due to a failure to resolve the replicated chromosomes. To circumvent this problem, we performed Hi-C on sporulating cells before and after inactivation of SMC. Sporulating cells contain two fully replicated chromosomes, and new rounds of replication are blocked by at least two distinct regulators (Rahn-Lee et al. 2009; Wagner et al. 2009; Boonstra et al. 2013). Prior to SMC inactivation, the long-range interactions within the origin region and between the chromosome arms were similar to wild-type sporulating cells (Fig. 3C; Supplemental Fig. S3G). However, 20 and 40 min after shifting to the restrictive temperature, these interactions were significantly reduced (Fig. 3C).
Figure 3.
Condensin is required for long-range chromosome interactions. (A) Normalized Hi-C contact matrices of a temperature-sensitive smc mutant (smcts; BWX3151) grown at 30°C or after 10, 20, and 40 min at 42°C. A wild-type strain (smc WT; BWX2080) grown at 30°C and shifted for 20 min to 42°C is shown for comparison. (B) Bar graph of the percentage of interarm interactions compared with total interactions for the Hi-C experiments in A and Figures 1 and 2 (see Supplemental Fig. S6A for details). (C) The smcts strain (BWX3266) induced to sporulate at 30°C and harvested after 190 min at 30°C (left), 170 min at 30°C and then 20 min at 42°C (middle), or 150 min at 30°C and then 40 min at 42°C (right). (D) Wild-type (PY79) and Δsmc (BDR2298) strains grown in minimal medium at 22°C.
As a final test, we performed Hi-C on wild-type and smc-null mutant cells grown in minimal medium at 22°C. Although all interaction probabilities appeared weaker under these conditions, the interarm interactions present in wild type were completely absent in the Δsmc mutant (Fig. 3D). Altogether, we conclude that condensin is required for the interactions between the two arms and between the origin and more distal sites. Our data further suggest that both types of long-range interactions require loading of condensin at the origin by ParB/parS nucleoprotein complexes.
Condensin complexes are present along the chromosome arms in sporulating cells
The requirement for condensin for interaction between the two chromosome arms raised the possibility that it acts not only at the origin but also along the length of the left and right arms. In support of this idea, ChIP-on-chip experiments indicate that SMC is enriched at origin-proximal parS sites but is also present at lower levels along the arms, forming a shallow enrichment gradient from origin to terminus (Gruber and Errington 2009). Cytological analysis of the condensin complex in exponentially growing cells indicates that SMC and ScpB colocalize with ParB in the origin region (Gruber and Errington 2009; Sullivan et al. 2009). Although localization of the condensin subunits along the chromosome arms is not readily apparent under these conditions, ScpB and SMC frequently form less discrete fluorescent foci than ParB, suggesting a more dispersed or extended localization (Supplemental Fig. S7A; Sullivan et al. 2009). We wondered whether we could detect condensin along the arms in sporulating cells where the chromosomes are more resolved because the replicated origins are tethered to opposite cell poles, and the remaining DNA forms an elongated structure that extends between them (Ben-Yehuda et al. 2003; Wang et al. 2014a). We visualized ScpB-YFP and TetR-CFP bound to an array of operators inserted adjacent to the origin at early stages of sporulation. As observed in vegetatively growing cells, fluorescent ScpB-YFP foci colocalized with the origin tag near the cell poles (Fig. 4A). However, weaker fluorescent foci were readily apparent along the DNA between them. Importantly, these ScpB foci were lost in cells lacking ParB (Fig. 4A), which is only present at the origin (Supplemental Fig. S7C; Sullivan et al. 2009). Immunoblot analysis indicates that ScpB-YFP, which localized as a diffuse haze, remained intact in the absence of ParB (Fig. 4B). Although it is possible that these results are specific to sporulation, they support the idea that condensin is loaded at the origin but is present and could act along the chromosome arms.
Figure 4.
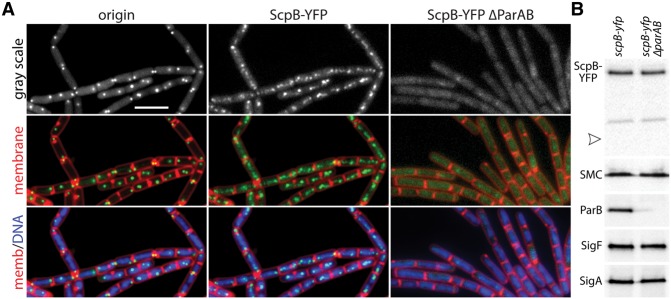
Condensin localizes along the chromosome arms during sporulation. (A) Representative fluorescence images of sporulating wild-type (BWX941) and ΔparAB (BWX3327) cells harboring an ScpB-YFP fusion. Cells were harvested 1 h after resuspension in sporulation medium. The replication origin was visualized using tetO/TetR-CFP, the membranes were stained with FM4-64, and the DNA was stained with DAPI. Bar, 4 µm. (B) Immunoblot analysis of the strains harboring ScpB-YFP in the presence (BKM1634) or absence (BWX3327) of ParAB. In the absence of ParAB, ScpB-YFP remained intact, and the level of SMC was similar to wild type. ScpB-YFP was detected using anti-GFP antibodies, and the arrowhead identifies the predicted size of free YFP. Both strains efficiently entered sporulation, as judged by the levels of the sporulation transcription factor SigF. SigA was used to control for loading.
A single origin-proximal parS site is sufficient for interarm interaction and specifies the inflection point
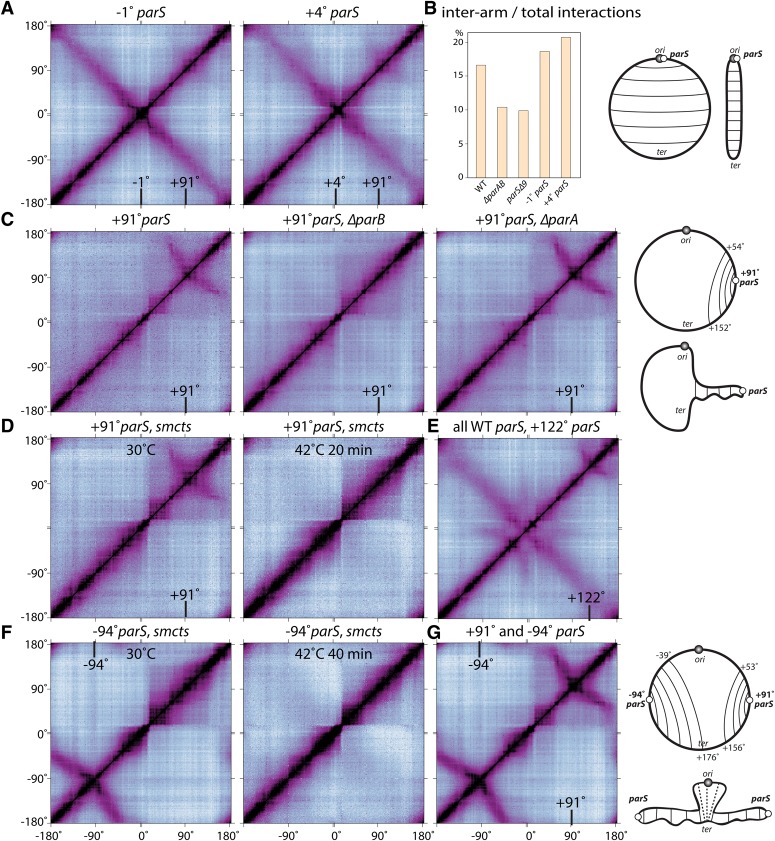
The B. subtilis chromosome has eight parS sites that are present in a 776-kb region around the origin (between −27° and +42°) and two origin-distal sites at +91° and −161°. All 10 sites are occupied by ParB in vivo as assayed by ChIP, although the origin-distal sites (especially the one at −161°) appear to be weaker than those present near the origin (Supplemental Fig. S5; Breier and Grossman 2007; Graham et al. 2014). Using a chromosome organization assay that takes advantage of polar division during sporulation, we showed previously that the organization of the origin region is perturbed in a strain lacking the eight origin-proximal parS sites (parSΔ8) but is largely restored by the insertion of a single parS site adjacent to the origin (Sullivan et al. 2009). Accordingly, we investigated whether a single origin-proximal parS site was sufficient to restore the long-range interactions observed by Hi-C. We performed Hi-C on a strain that contained the endogenous parS site at −1° (10 kb from the origin) but lacked the other seven origin-proximal sites. Strikingly, interarm interactions along the entire left and right chromosome arms were retained, while interactions between the origin region and more distal sites were absent (cf. Figs. 5A and 1A). Quantification of interaction frequencies between the two arms revealed stronger and more spatially restricted interactions in this strain, as if the arms were juxtaposed more tightly or at more regular intervals than in wild type (Fig. 5B; Supplemental Fig. S6C). To investigate whether other endogenous parS sites behave similarly, we performed Hi-C on a strain that contained a single parS site at +4° (48 kb from the origin). Again, interarm interactions were retained, but long-range interactions between the origin and distal sites were absent (Fig. 5A,B; Supplemental Fig. S6C). Importantly, in both cases, the inflection point for the alignment of the two arms closely correlated with the position of the parS site (Supplemental Fig. S6C). Collectively, these data indicate that a single origin-proximal parS site is both necessary and sufficient to promote interarm interactions along the entire length of the chromosome.
Figure 5.
Alignment of DNA flanking parS sites. (A) Normalized Hi-C contact matrices of strains with a single origin-proximal parS site at −1° (BDR2996) or +4° (BDR2985). Both strains also contain the weak parS site at +91°. A bar graph of the percentage of interarm interactions compared with total interactions for the indicated strains is shown in B (see Supplemental Fig. S6A for details). (C) Strains harboring the endogenous parS site at +91° but lacking all eight origin-proximal parS sites (parSΔ8) in an otherwise wild-type background (BNS1657) or in cells lacking parB (BWX2761) or parA (BDR3007). (D) A strain (BWX3304) harboring the endogenous parS site at +91° and smcts grown at 30°C and shifted for 20 min to 42°C. (E) A strain (BWX3172) with an ectopic parS site inserted at +122° in an otherwise wild-type background. (F) A strain (BWX3359) harboring an ectopic parS site at −94° and smcts grown at 30°C and shifted for 40 min to 42°C. A 20-min time point was not tested. (G) A strain (BWX3231) with two parS sites inserted to +91° and −94° in parSΔ9. The positions of the relevant parS sites (in degrees) are indicated in the contact maps. Schematic representations of the long-range interactions and the disposition of the chromosome are shown at the right (see also Supplemental Fig. S8A,B).
Origin-distal parS sites promote juxtaposition of flanking DNA
Next, we investigated whether an origin-distal parS site could promote interarm interactions. To do this, we performed Hi-C on the strain lacking all eight origin-proximal parS sites (parSΔ8) but harboring the weak origin-distal sites at +91° and −161°. The resulting contact matrix lacked interarm interactions (the secondary diagonal) but revealed new long-range interactions that were centered on the +91° parS site (Fig. 5C; Supplemental Fig. S8A). DNA on either side of this site was linearly juxtaposed over a large portion (∼1.1 Mb) of the right arm. Specifically, 418 kb origin-proximal to the parS site (from +54° to +91°) and 682 kb terminus-proximal to the parS site (from +91° to +152°) appeared to have “zipped up” with the +91° parS site at the inflection point. This novel conformation (referred to as a zip-up for simplicity) required the parS site at +91° (Fig. 2A), ParB (Fig. 5C), and SMC (Fig. 5D) but not ParA (Fig. 5C), consistent with the idea that these interactions are similarly promoted by condensin loaded at ParB/parS.
We suspected that this unusual interaction signature, which was not observed in the contact matrix from wild-type cells (Fig. 1A), was due to the strength of the parS site at +91°. This site binds significantly less ParB than the sites in the origin region, as assayed by ChIP (Supplemental Fig. S5; Breier and Grossman 2007). In the absence of the eight origin-proximal parS sites, ParB occupancy at this weaker site likely increases, resulting in more recruitment of condensin. Consistent with this idea, weak zip-up interactions centered at +91° were apparent in the strains that harbor a single origin-proximal parS site (cf. Figs. 5A and 2A). To more directly test this, we performed Hi-C on a wild-type strain in which a strong parS site was inserted at +122°. In addition to the long-range interactions normally present in wild-type cells, interactions between DNA flanking the +122° parS site were observed (Fig. 5E).
Finally, to investigate the consequences of a strong origin-distal parS site in a strain lacking the native sites, we performed Hi-C on parSΔ9 with the consensus parS site from +4° (Breier and Grossman 2007) transplanted to −94°. Similar condensin-dependent zip-up interactions were observed (Fig. 5F; Supplemental Fig. S8A). These long-range interactions were even more pronounced than the parSΔ8 strain harboring the endogenous +91° parS site. Six-hundred-ten kilobases origin-proximal to the ectopic parS site (from −39° to −94°) and 1013 kb terminus-proximal (from −94° down to +176°) were aligned with the inflection point at the parS site (at −94°). As in the case with the endogenous parS site at +91°, the DNA flanking the −94° parS site was not aligned symmetrically. Approximately 1.7 times more DNA terminus-proximal to the parS site was aligned with origin-proximal DNA (Supplemental Fig. S8A). One explanation for this asymmetry is suggested by the origin-proximal boundaries for the zip-up interactions generated by the parS sites at +91° and −94°. Both overlap highly transcribed operons (Nicolas et al. 2012) that are co-oriented with replication. Specifically, the rrnE locus (at +53°) and the operon encoding ATP synthase (atpI to atpC at −39°) appeared to attenuate further zip-up interactions (Supplemental Fig. S8A). Intriguingly, in B. subtilis, 74% of all genes and 87% of the 400 most highly transcribed genes in rich medium are present on the leading strand (Rocha 2002; Nicolas et al. 2012). Thus, if the alignment of DNA flanking parS sites is indeed influenced by the orientation of transcription units, this could explain the asymmetric zip-up interactions generated by the −91° and +94° parS sites and also the symmetric and complete alignment of the two arms in the strains harboring a single origin-proximal parS site. It could also provide an explanation for why the zip-up interactions did not extend significantly beyond the replication terminus.
Origin-distal parS sites act independently
Next, we investigated whether parS sites on opposing arms act independently or together to structure the chromosome. We inserted identical parS sites (from +4°) at +91° and −94° in the parSΔ9 strain and analyzed the conformation of the chromosome by Hi-C. Each parS site promoted independent long-range interactions, zipping up flanking DNA (Fig. 5G; Supplemental Fig. S8A,B). These results argue that the factors that generate these interactions act along adjacent DNA segments rather than between DNA on opposing sides of the chromosome.
This analysis also allowed us to directly compare the interactions generated by a weak (endogenous) parS site at +91° (Fig. 5C; Supplemental Fig. S8A) with those generated by a strong ectopic parS site (Fig. 5G; Supplemental Fig. S8A) at the same position. Although the stronger parS site promoted interactions between somewhat larger regions of DNA (1.2 Mb compared with 1.1 Mb), the alignment of the origin-proximal DNA did not extend beyond the CID bounded by the rrnE locus, and the alignment as a whole was similar, as can be seen by the shape of the interaction signature in the contact matrices. Finally, similar to what was observed in strains with a single origin-distal parS, the alignment of DNA flanking the +91° and −94° parS sites did not extend beyond the terminus region. Instead, the terminus interacted with a broad region of DNA surrounding the origin that included two large CIDs flanked by highly transcribed genes (Supplemental Fig. S8B).
Origin-distal parS sites bring flanking loci into close proximity
To assess whether the juxtaposition of flanking DNA observed by Hi-C reflects the disposition of the chromosome in live cells, we examined the distance between loci on either side of origin-distal parS sites. We inserted tetO and lacO arrays at −64° and −125° into the parSΔ9 strain and monitored their localization in the presence and absence of an ectopic parS site at −94°. We collected snapshot images of cells grown in rich and minimal media and analyzed the distance between pairs of loci (see the Supplemental Material). Interfocal distances decreased significantly when the ectopic parS site was present during fast and slow growth (Supplemental Fig. S8C). A similar parS-dependent reduction in interfocal distances was observed when we monitored loci at +54° and +135° in the presence and absence of the endogenous parS site at +91° (Supplemental Fig. S8D). These experiments provide independent confirmation that origin-distal parS sites can promote juxtaposition of large tracks of flanking DNA.
Condensin recruitment to origin-proximal parS sites structures the origin region
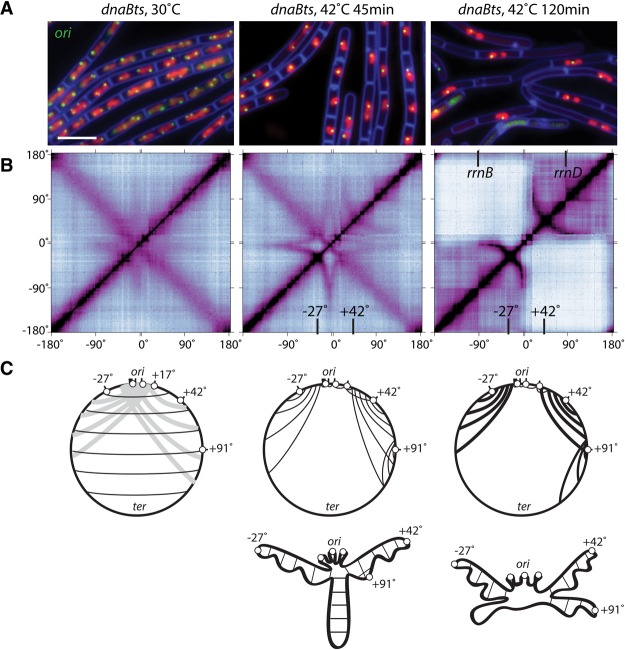
The contact probability matrices of wild-type cells displayed unusual but poorly resolved long-range interaction signatures between the origin region and distal sites (Fig. 1A; Supplemental Fig. S3A). These interactions required ParB (Fig. 2A), the origin-proximal parS sites (Fig. 2A), and condensin complexes (Fig. 3). Furthermore, a single parS was not sufficient to restore them (Fig. 5A). Our data indicating that origin-distal parS sites promote zip-up of flanking DNA raised the possibility that the interaction signatures in the origin region might result from similar alignment of DNA flanking each of the eight origin-proximal parS sites. We wondered whether these interactions would be more resolved in the absence of ongoing replication, which likely disrupts these structures as the replication forks pass through them. Accordingly, we performed Hi-C on a temperature-sensitive replication initiation mutant (dnaBts) (Rokop et al. 2004) under permissive conditions (30°C) and after shifting to the restrictive temperature (42°C) to generate cells with a single nonreplicating chromosome. Based on fluorescence imaging, after 45 min at 42°C, most cells had completed DNA replication and contained a spherical nucleoid with a single replication origin (Fig. 6A; Supplemental Fig. S9A). Furthermore, by 120 min, the nucleoid resolved into a bilobed structure with the origin near the center and the left and right arms in separate lobes (Fig. 6A; Supplemental Fig. S9A; Wang et al. 2014a). The interaction matrix generated from dnaBts cells grown at 30°C looked similar to wild type (cf. Figs. 6B and 1A; Supplemental Fig. S4B). In contrast, after 45 min at 42°C, the contact map contained well-defined long-range interactions in the origin region (Fig. 6B; Supplemental Fig. S9B). The most clearly resolved interactions emanated from the parS sites at −27° and +42° in which DNA flanking these sites was juxtaposed. Similar to the origin-distal sites (Fig. 5C–G), significantly more DNA from the terminus-proximal side of these parS sites interacted with origin-proximal DNA, generating the signature arcs in the contact matrix. The parS sites that reside between −27° and +42° (−6°, −5°, −4°, −1°, +4°, and +17°) appeared to promote similar zip-up interactions but were more difficult to resolve. We suspect that each parS site competes for flanking DNA, resulting in smaller juxtaposed regions. In addition, the highly transcribed ribosomal RNA loci (rrnO, rrnA, rrnJW, and rrnIHG between 0.9° and 15°) and the large operon encoding ribosomal proteins and RNA polymerase (between 11° and 14°) may further affect the ability to detect these interactions (Supplemental Fig. S9B). Nonetheless, based on the similarities between the contact matrix from the asynchronous cultures and the one from the replication-arrested cells, we hypothesize that condensin complexes recruited by ParB at each parS promote the formation of independently juxtaposed domains.
Figure 6.
Long-range interactions emanating from the origin are revealed in cells blocked for replication initiation. (A) Representative fluorescence images of cells (BWX2533) harboring a temperature-sensitive replication initiation mutant in dnaB (dnaBts) at permissive (30°C) and restrictive (42°C) temperatures for the indicated times. The replication origins (ori) were visualized using tetO/TetR-CFP (green), the DNA was visualized with DAPI (red), and the cell membranes were visualized with FM4-64 (blue). Bar, 4 µm. (B) Normalized Hi-C contact matrices for the dnaBts strain (BNS1733) under the same conditions as in A. The locations of relevant parS sites at −27° and +42° and rrn loci are indicated (see also Supplemental Fig. S9). (C) Schematic representations of the long-range interactions and the disposition of the chromosome under the different conditions. The nine parS sites (black bars) are represented by six white balls: The cluster of −1°, −4°, −5°, and −6° parS sites are represented by a single ball, and the +4°, +17°, +42°, +91°, and −27° parS sites are represented by five separate balls. (Left panel) Light-gray lines in the schematic of cells grown under the permissive condition highlight the poorly resolved long-range interactions between the origin and more distal sites. (Middle panel) For simplicity, the interarm interactions are not shown, and only the zip-up interactions at the parS sites are presented. (Right panel) Thicker lines represent stronger interactions in the contact map.
After 120 min at the restrictive temperature, the contact map changed dramatically (Fig. 6B). Interactions between the two arms were completely absent, and the chromosome resolved into two independent domains, each corresponding to a replication arm. This conformation matches previous cytological analysis (Wang et al. 2014a) and provides further validation for our Hi-C results. In addition, a 220-kb region spanning the terminus, which is bounded by the highly transcribed glnRA and odhAB operons and contains the dif site, formed a domain that had contact with the entire chromosome (Supplemental Fig. S9C), consistent with the idea that this region connects the two lobes (Wang et al. 2014a). Finally, long-range interactions at sites close to the origin were absent and were replaced by short-range interactions that overlapped the highly transcribed rrn loci at most origin-proximal parS sites (Supplemental Fig. S9B). Within the domains made up of the left and right chromosome arms, the parS sites at −27° and +42° promoted alignment of flanking DNA (Fig. 6B; Supplemental Fig. S9B). These interactions were significantly stronger than at the 45-min time point. Furthermore, the DNA that is zipped up on the origin-proximal side of these parS sites appeared to be attenuated by the CIDs that contain the rrn loci adjacent to the origin. Altogether, these results suggest that the long-range interactions emanating from the origin in actively replicating cultures result from the juxtaposition of DNA flanking the origin-proximal parS sites and that these interactions require ParB/parS-loaded condensin.
Discussion
In toto, this study provides a more complete picture of how the B. subtilis chromosome is organized, establishes the central role played by condensin (and its recruitment by ParB to parS sites) in structuring the genome, and provides insight into how condensin complexes resolve newly replicated origins.
Previous cytological analysis revealed that, for most of the replication-segregation cycle, the B. subtilis chromosome resides in an ori-ter configuration with left and right arms aligned side by side (Wang et al. 2014a). The Hi-C experiments presented here provide confirmation of this organization and argue that the two arms reside in close proximity along their entire lengths. Importantly, these long-range interactions were undetectable in cells lacking ParB, the origin-proximal parS sites, or the condensin complex. The loss of interarm interactions in these mutants could reflect a subtle defect in alignment in which the two arms lose register, and the interactions between them span a broader region. Alternatively, it could result from a conformation in which the two arms are organized into separate entities. The absence of interarm interactions in cells lacking ParB, the parS sites, and SMC (Figs. 1–3) and the formation of CIDs that span each arm favor a model in which these factors indirectly or directly promote the interaction between the two arms, and, in their absence, the arms become physically separate. Further support for this idea comes from our findings that the distance between chromosomal loci on opposite arms increased in cells lacking ParAB (but not ParA) (Fig. 1C; Supplemental Fig. S3C,D,F), and origin-distal parS sites promote the alignment of flanking DNA.
Our Hi-C contact matrices also revealed a set of long-range interactions between the origin region and more distal sites. These interactions similarly depended on ParB, the origin-proximal parS sites, and condensin, suggesting that they too are mediated by condensin action on either side of the parS sites in the origin region. In support of this idea, cells blocked for replication initiation have similar but more clearly resolved interaction patterns in which DNA flanking origin-proximal parS sites is juxtaposed. We propose that the origin region adopts a conformation in which DNA flanking each parS site (or possibly a cluster of sites, in the case of −4°, −5°, and −6°) aligns into finger-like projections (Fig. 6C). Each origin-proximal parS site likely competes for flanking DNA such that the sites closest to the origin align short stretches of DNA, while the two more distal sites (at −27° and +42°) have access to large expanses of terminus-proximal DNA and generate the long-range interactions observed in our contact matrices. Our observation that cells with a single parS site retain interarm interactions but lose the long-range interactions at the origin is fully consistent with this model.
We note that the contact matrix from cells inhibited for replication initiation for 45 min had an interaction arc that emanated from the secondary diagonal at −27° (Supplemental Fig. S9B, black arrows). We suspect that the interactions that generated it arise when condensin loads at the −27° parS site and promotes alignment of flanking DNA. Since this DNA is itself juxtaposed with DNA on the opposing arm, the opposing DNA becomes incorporated into the same zipped-up structure. Future experiments will be aimed at better understanding these unusual long-range interactions.
We and others have previously shown that origin-localized condensin complexes play a critical role in individualizing replicated origins in B. subtilis (Gruber et al. 2014; Wang et al. 2014b). We proposed that condensin resolves the origins by compacting newly replicated DNA along its length, drawing it in on itself and away from its sister origin. At that time, we envisioned condensin constraining individual loops of DNA adjacent to parS sites. However, based on the data presented here, we suggest that condensin draws sister origins apart by lengthwise compaction but does so by promoting juxtaposition of DNA flanking origin-proximal parS sites (Fig. 7D). In eukaryotes, sites of condensin enrichment have been identified in a few model organisms by ChIP (D'Ambrosio et al. 2008; Kim et al. 2013; Kranz et al. 2013; Nakazawa et al. 2015) and appear to be scattered throughout the chromosomes. If these represent loading sites that function similarly to ParB/parS, then alignment of flanking DNA could provide a mechanism to promote compaction and resolution of chromosomes into discrete bodies during the early stages of mitosis.
Figure 7.
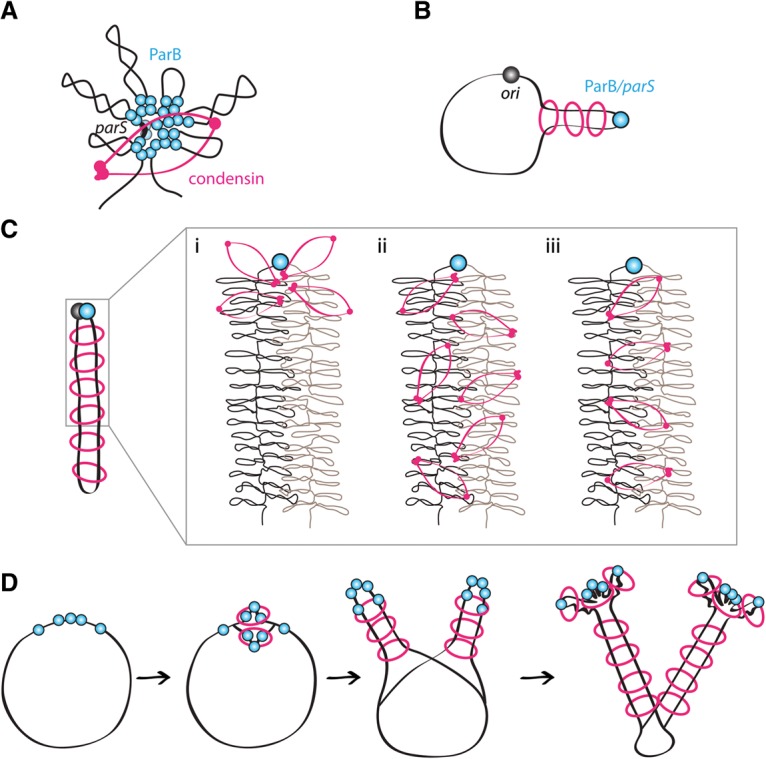
Condensin rings tether DNA flanking parS sites and promote resolution of sister origins. (A) Schematic models of condensin (pink) loading at a ParB/parS nucleoprotein complex (light blue) (Graham et al. 2014; Wilhelm et al. 2015). (B) Condensin loaded at origin-distal parS sites aligns flanking DNA. (C) Condensin loaded at a single origin-proximal parS site juxtaposes the two arms. The insert shows three models for condensin action. (Panel i) Condensin acts adjacent to the parS site, setting up a compacted state that is propagated down the flanking DNA. The two condensed chromosomal arms are brought into close proximity by other proteins or due to confinement and/or crowding. (Panel ii) Condensin acts along the two arms to compact them and indirectly promotes their interaction. (Panel iii) Condensin loaded at parS encircles the flanking DNA strands. Migration down the DNA tethers the two strands of DNA. The supercoiled loops are shown schematically and are several times larger than depicted. (D) Condensin loading at newly replicated origin-proximal parS sites draws sister origins in on themselves and away from each other. Alignment of DNA flanking parS sites at −27° and +42° may impede movement of origin-loaded condensin rings down the arms. Although condensin rings are depicted encircling two strands of DNA in these models, the same activities would result from separate rings encircling DNA on either side of the loading site and migrating as unit (handcuffed) down the tethered DNA (see Supplemental Fig. S10).
Unlike the smcts mutant, cells lacking ParB are impaired but not blocked in origin segregation and have a relatively modest defect in chromosome segregation (Lee and Grossman 2006; Wang et al. 2014b). However, in its absence, long-range interactions along the arms and between the origin and more distal sites were virtually undetectable by Hi-C (Figs. 1A, 2A). These results raise the possibility that zip-up interactions may not be critical for origin segregation. However, recent work indicates that a small proportion of condensin complexes is able to encircle the chromosome in the absence of ParB (Wilhelm et al. 2015). Accordingly, we suspect that this small number of complexes is sufficient to resolve origins by promoting alignment of DNA in a manner independent of ParB/parS but is unable to generate detectable long-range interactions in the population-based Hi-C assay.
Models for condensin action
We envision three possible models for how condensin promotes juxtaposition of flanking DNA. In the first, origin-loaded condensin constrains DNA segments in the origin region, and this organization is somehow propagated down the two chromosome arms independently of condensin resulting in their apposition (Fig. 7C, panel i). In the second, condensin loaded at the origin acts along the two chromosome arms flanking its loading site and then indirectly or directly brings the compacted structures together (Fig. 7C, panel ii). In the third model, condensin is loaded onto DNA by ParB such that the ring encircles two DNA helices: one from each side of the parS site (Fig. 7A). As the ring slides away from the ParB/parS complex, it tethers the flanking DNA strands together (Fig. 7C, panel iii). Alternatively, two interacting or interlocked condensin rings could topologically entrap DNA on either side of the parS site, and these “handcuffs” (Thadani et al. 2012) would migrate down the flanking DNA (Supplemental Fig. S10).
Although we cannot rule out the first two models (Fig. 7C, panels i,ii), we favor the “loop extrusion” models (Fig. 7C, panel iii; Supplemental Fig. S10) originally proposed by Nasmyth (2001) and more recently elaborated by Alipour and Marko (2012) because they provide a simple explanation for how parS sites could serve as the inflection point for DNA juxtaposition (Fig. 5; Supplemental Fig. S8A,B), most easily explain how ectopic parS sites could bring flanking DNA into juxtaposition (Figs. 5, 7B; Supplemental Fig. S8A,B), and explain how alignment occurs specifically along contiguous DNA and not between parS sites on opposing arms (Fig. 5G; Supplemental Fig. S8A,B). Furthermore, they can account for the increased frequency of interarm interactions in cells with a single origin-proximal parS site (Fig. 5B; Supplemental Fig. S6C) and the increased intra-arm interactions on either side of −27° and +42° parS sites at 120 min after replication arrest (Fig. 6B; see below; Supplemental Fig. S9B). These models are also consistent with the ability of condensin to entrap DNA in vivo (Wilhelm et al. 2015). The size of the pore in the condensin ring (>35 nm in the largest dimension) (Burmann et al. 2013) could easily accommodate one or two DNA strands with attendant DNA-binding proteins, and supercoiled loops are likely flexible and dynamic enough to not obstruct movement (Fig. 7C). Finally, this model ensures that condensin acts on contiguous DNA segments, driving intrachromosomal compaction while avoiding unproductive interchromosomal entanglement (Marko 2009).
Our data suggesting that the orientation of transcription influences DNA alignment can be understood in the context of these loop extrusion models. When condensin is loaded at the origin, co-oriented transcription could facilitate movement by buffeting the rings down the two arms. When loaded at origin-distal sites, transcription on the leading strand would slow movement of origin-proximal DNA through the ring while promoting the movement of terminus-proximal DNA, resulting in the observed asymmetric juxtaposition. In the most extreme situation, a large highly transcribed operon origin-proximal to the parS site could completely prevent migration. A similar model for the movement of cohesin complexes away from their loading sites has been proposed based on their accumulation at intergenic regions between convergently transcribed loci (Ocampo-Hafalla and Uhlmann 2011).
We note that the mechanism by which condensin moves down the chromosome arms cannot be explained by our data and serves as an important challenge to the loop extrusion models. However, the mechanism by which juxtaposition of flanking DNA is propagated away from parS sites is unclear in all models. It has been suggested that the energy for ring movement could be provided by co-oriented transcription (Gruber 2014). However, we showed here that ectopic parS sites juxtaposed large tracks of origin-proximal DNA in which ∼74% of the genes are present on the leading strand. Furthermore, in the strain harboring a single parS site at +91°, alignment of flanking DNA extended beyond the convergently transcribed rrnD locus (at +81°) 110 kb away (Fig. 5C; Supplemental Fig. S8A). Similarly, the parS site at −94° promoted juxtaposition well beyond the rrnB operon (at −93°) 11 kb away (Fig. 5F,G; Supplemental Fig. S8A). The alignment of DNA in these regions was highly asymmetric, suggesting that these operons could impede movement but clearly do not prevent it. Although our data do not bear on this issue, one possibility is that condensin's ATPase activity could provide the energy for ring movement (Strick et al. 2004; Alipour and Marko 2012). In this model, ParB/parS would load condensin onto the DNA strands in an orientation that would allow processive movement away from parS. Our data indicate that productive recruitment (and likely loading) of condensin by ParB requires both the ParB/parS nucleoprotein complex (Fig. 2A) and interaction between condensin and its cognate ParB (Fig. 2B–E). However, the nature of these interactions and the mechanism and possible orientation of condensin loading await future biochemical and structural studies.
Condensin-mediated interarm interactions
One apparent discrepancy between our data and the proposed model relates to condensin-dependent interarm interactions in wild-type cells. In the case where cells were engineered to contain a single origin-proximal parS site, condensin loading at this site would tether the two arms, and all of the rings could move down the arms unimpeded, generating strong and spatially restricted interarm interactions (Fig. 7C). However, in wild-type cells with eight origin-proximal parS sites, condensin-mediated zip-up of DNA flanking the −27° and +42° parS sites could effectively serve as barriers to movement of the rings loaded at the origin that tether the two arms (Fig. 6C). We hypothesize that condensin rings that are loaded at the parS sites adjacent to the origin generate interarm tethers shortly after their replication and migrate down the arms before condensin loaded at the −27° and +42° sites accumulates (Fig. 7D). Once there is sufficient condensin-mediated alignment of DNA flanking these more distal sites, replenishment of interarm tethers would require a new round of replication. Consistent with this model, condensin (as assayed by ScpB-GFP) was less concentrated in the origin region in the strain harboring a single origin-proximal parS site than in wild type (Supplemental Fig. S7B), suggesting that more complexes were able to traverse the two arms. Furthermore, this model predicts that, if replication initiation is blocked, the condensin rings that align the two arms will not be replenished, and those that were loaded in the last round of replication would be lost by migration to the terminus and/or release from the chromosome. In contrast, additional condensin loaded at the −27° and +42° parS sites would further reinforce the alignment of DNA flanking these sites. These predictions are borne out 120 min after blocking replication initiation (Fig. 6B). At this time point, all interaction between the two arms is lost, and the interactions flanking −27° and +42° are stronger. We hypothesize that the strong intra-arm interactions at these parS sites and the absence of interarm tethers drive the resolution of the two arms and the formation of the bilobed structure. We showed previously that, during an unperturbed replication–segregation cycle, shortly after replication initiation, the replicated origins move to midcell, and the template DNA adopts a left-ori/ori-right configuration (Wang et al. 2014a). In principal, this transition could be similarly driven by increased interactions flanking the −27° and +42° parS sites due to release of origin-proximal condensin upon replication initiation.
In conclusion, juxtaposition of DNA flanking parS sites mediated directly or indirectly by condensin provides a mechanism for driving intrachromosomal compaction and origin resolution while avoiding unproductive interchromosomal entanglement. Establishing how condensin promotes these interactions is the central challenge for the future.
Materials and methods
General methods
B. subtilis strains were derived from the prototrophic strain PY79 (Youngman et al. 1983). Cells were grown in defined rich (CH) (Harwood and Cutting 1990) or minimal (S750) (Grossman and Losick 1988) medium supplemented with 1% sorbitol. Sporulation was induced by resuspension according to the method of Sterlini-Mandelstam (Harwood and Cutting 1990). For Hi-C experiments during sporulation, strains contained the SpoIIIE DNA translocase mutant (spoIIIE36) that engages the chromosome after polar division but does not transport DNA (Wu and Errington 1994). Lists of strains, plasmids, and descriptions of their construction are in the Supplemental Material.
Hi-C
The Hi-C protocol was adapted from Le et al. (2013). Briefly, cells were cross-linked with 3% formaldehyde for 30 min at room temperature, quenched with 125 mM glycine for 5 min, washed with 1× PBS buffer, resuspended in 1× PBS at a final concentration of 1 × 107 cells per microliter, and frozen in 50-µL aliquots. For each Hi-C reaction, 5 × 107 cells were used. One frozen aliquot (10 reaction equivalents), was thawed, pelleted, resuspended in 50 µL of TE containing ready-lyse lysozyme (Epicentre, R1802M) and protease inhibitors (Sigma, P8340), and incubated for 30 min at room temperature. Ten percent SDS was then added to a final concentration of 0.5% and incubated for 10 min at room temperature. Five microliters of solubilized chromatin was digested using 2.5 µL of 250 U/µL HindIII for 30 min at 37°C. The cleaved ends were filled in with Klenow and biotin-14-dATP, dGTP, dCTP, and dTTP (45 min at 20°C) followed by the addition of SDS to 0.5%. The products were then ligated in 1-mL volume with 3 µL of 2000 U/µL T4 DNA ligase overnight at 16°C. Cross-linking was reversed overnight at 65°C in the presence of 5 µL of 0.8 U/µL proteinase K. The DNA was then extracted twice with phenol/chloroform/isoamyl alcohol (PCI; 25:24:1), precipitated with ethanol, and resuspended in 20 µL of Qiagen EB buffer. Biotin from nonligated ends was removed using T4 polymerase (4 h at 20°C) followed by extraction with PCI. The DNA was then sheared by sonication in 10 sec on–10 sec off cycles for 12 min with 60% amplitude using a Qsonica (Q800) water bath sonicator. The sheared DNA was used for library preparation with the NEBNext Ultra kit (E7370S) according to the manufacturer's instructions for end repair, adapter ligation, and size selection. Biotinylated DNA fragments were purified using streptavidin beads as in Le et al. (2013) except that 10 µL of beads was used for each sample, and beads were resuspended in 10 µL of ddH2O. Five microliters of DNA-bound beads was used for PCR in a 50-µL reaction for 14 cycles. PCR products were purified using Ampure beads and sequenced at the Tufts University Core Facility using HiSeq 2500. Paired-end sequencing reads were mapped to the genome of B. subtilis PY79 (NCBI reference sequence NC_022898.1). A detailed description of Hi-C mapping and analysis is in the Supplemental Material. The genetic loci marked by degrees were calculated using the PY79 genome, which results in a slight shift from previously published data using B. subtilis strain 168 genomic coordinates.
Fluorescence microscopy
Fluorescence microscopy was performed on a Nikon Ti microscope equipped with a plan apo 100×/1.4 NA phase-contrast oil objective and a CoolSnapHQ2 camera. Cells were immobilized using 2% agarose pads containing growth medium. Membranes were stained with FM4-64 (Molecular Probes) at 3 μg/mL. DNA was stained with DAPI at 2 μg/mL. Images were cropped and adjusted using MetaMorph software (Molecular Devices). Final figures were prepared in Adobe Illustrator. Image analysis was performed using MicrobeTracker (Sliusarenko et al. 2011); details are in the Supplemental Material.
ChIP-seq
ChIP was performed as described previously (Graham et al. 2014) with the following modifications. Cells were grown in defined rich (CH) medium to an optical density of 0.5 and cross-linked using 3% formaldehyde for 30 min at room temperature. Cells were then quenched, washed, and lysed as described previously (Graham et al. 2014). Chromosomal DNA was sheared to an average size of 200 base pairs by sonication using a Qsonica (Q800) water bath sonicator. The lysate was then incubated overnight at 4°C with anti-mCherry antibodies. The sample was then processed as described previously and sequenced using the Illumina MiSeq platform. The sequencing reads were aligned to the B. subtilis PY79 genome (NC_022898.1) using CLC Genomics Workbench software. Anti-mCherry antiserum was generated using recombinant (His)6-mCherry expressed in and purified from Escherichia coli BL21 DE3 pLysS. Peak fractions from Ni2+ affinity chromatography were used to generate polyclonal antibodies in rabbits (Covance).
Accession numbers
All sequence data reported in this study have been deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) with accession number GSE68418.
Supplementary Material
Acknowledgments
We thank members of the Rudner and Bernhardt laboratories for stimulating discussions and support, Paula Montero-Llopis and Stephan Uphoff for help with Matlab scripts, Brendan Taillon for strain construction, John Marko for insights, the referees for suggesting additional controls, and the Tufts University Core Facility for their assistance with high-throughput sequencing. A special thank you to Marcelo Nollmann and coworkers for open communication throughout this research. Support for this work comes from National Institutes of Health grants (GM086466 and GM073831 to D.Z.R., and GM082899 to M.T.L.) and National Human Genome Research Institute grants (HG003143 and HG007010 to J.D.). X.W. was a long-term fellow of the Human Frontier Science Program. T.B.K.L is a Gordon and Betty Moore Foundation post-doctoral fellow of the Life Sciences Research Foundation. M.T.L. is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.265876.115.
References
- Alipour E, Marko JF. 2012. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res 40: 11202–11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299: 532–536. [DOI] [PubMed] [Google Scholar]
- Boonstra M, de Jong IG, Scholefield G, Murray H, Kuipers OP, Veening JW. 2013. Spo0A regulates chromosome copy number during sporulation by directly binding to the origin of replication in Bacillus subtilis. Mol Microbiol 87: 925–938. [DOI] [PubMed] [Google Scholar]
- Breier AM, Grossman AD. 2007. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol Microbiol 64: 703–718. [DOI] [PubMed] [Google Scholar]
- Britton RA, Lin DC, Grossman AD. 1998. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev 12: 1254–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmann F, Shin HC, Basquin J, Soh YM, Gimenez-Oya V, Kim YG, Oh BH, Gruber S. 2013. An asymmetric SMC–kleisin bridge in prokaryotic condensin. Nat Struct Mol Biol 20: 371–379. [DOI] [PubMed] [Google Scholar]
- Cuylen S, Haering CH. 2011. Deciphering condensin action during chromosome segregation. Trends Cell Biol 21: 552–559. [DOI] [PubMed] [Google Scholar]
- Cuylen S, Metz J, Haering CH. 2011. Condensin structures chromosomal DNA through topological links. Nat Struct Mol Biol 18: 894–901. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. 2008. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev 22: 2215–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TG, Wang X, Song D, Etson CM, van Oijen AM, Rudner DZ, Loparo JJ. 2014. ParB spreading requires DNA bridging. Genes Dev 28: 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AD, Losick R. 1988. Extracellular control of spore formation in Bacillus subtilis. Proc Natl Acad Sci 85: 4369–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S. 2014. Multilayer chromosome organization through DNA bending, bridging and extrusion. Curr Opin Microbiol 22: 102–110. [DOI] [PubMed] [Google Scholar]
- Gruber S, Errington J. 2009. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137: 685–696. [DOI] [PubMed] [Google Scholar]
- Gruber S, Veening JW, Bach J, Blettinger M, Bramkamp M, Errington J. 2014. Interlinked sister chromosomes arise in the absence of condensin during fast replication in B. subtilis. Curr Biol 24: 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. Wiley, New York. [Google Scholar]
- Hirano T. 2012. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev 26: 1659–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. 1994. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 79: 449–458. [DOI] [PubMed] [Google Scholar]
- Hirano M, Anderson DE, Erickson HP, Hirano T. 2001. Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J 20: 3238–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K, Gunther NW, Grossman AD. 1994. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol 176: 5320–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada K, Miyata M, Hirano T. 2013. Molecular basis of SMC ATPase activation: role of internal structural changes of the regulatory subcomplex ScpAB. Structure 21: 581–594. [DOI] [PubMed] [Google Scholar]
- Kim JH, Zhang T, Wong NC, Davidson N, Maksimovic J, Oshlack A, Earnshaw WC, Kalitsis P, Hudson DF. 2013. Condensin I associates with structural and gene regulatory regions in vertebrate chromosomes. Nat Commun 4: 2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hirano T. 1997. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90: 625–634. [DOI] [PubMed] [Google Scholar]
- Kranz AL, Jiao CY, Winterkorn LH, Albritton SE, Kramer M, Ercan S. 2013. Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol 14: R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TB, Imakaev MV, Mirny LA, Laub MT. 2013. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Grossman AD. 2006. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol Microbiol 60: 853–869. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Grossman AD. 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92: 675–685. [DOI] [PubMed] [Google Scholar]
- Livny J, Yamaichi Y, Waldor MK. 2007. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J Bacteriol 189: 8693–8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbouty M, Cournac A, Flot JF, Marie-Nelly H, Mozziconacci J, Koszul R. 2014. Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms. Elife 3: e03318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko JF. 2009. Linking topology of tethered polymer rings with applications to chromosome segregation and estimation of the knotting length. Phys Rev E Stat Nonlin Soft Matter Phys 79: 051905. [DOI] [PubMed] [Google Scholar]
- Mascarenhas J, Soppa J, Strunnikov AV, Graumann PL. 2002. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J 21: 3108–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray H, Ferreira H, Errington J. 2006. The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol Microbiol 61: 1352–1361. [DOI] [PubMed] [Google Scholar]
- Nakazawa N, Sajiki K, Xu X, Villar-Briones A, Arakawa O, Yanagida M. 2015. RNA pol II transcript abundance controls condensin accumulation at mitotically up-regulated and heat-shock-inducible genes in fission yeast. Genes Cells 20: 481–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35: 673–745. [DOI] [PubMed] [Google Scholar]
- Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, et al. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335: 1103–1106. [DOI] [PubMed] [Google Scholar]
- Ocampo-Hafalla MT, Uhlmann F. 2011. Cohesin loading and sliding. J Cell Sci 124: 685–691. [DOI] [PubMed] [Google Scholar]
- Petrushenko ZM, Cui Y, She W, Rybenkov VV. 2010. Mechanics of DNA bridging by bacterial condensin MukBEF in vitro and in singulo. EMBO J 29: 1126–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn-Lee L, Gorbatyuk B, Skovgaard O, Losick R. 2009. The conserved sporulation protein YneE inhibits DNA replication in Bacillus subtilis. J Bacteriol 191: 3736–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E. 2002. Is there a role for replication fork asymmetry in the distribution of genes in bacterial genomes? Trends Microbiol 10: 393–395. [DOI] [PubMed] [Google Scholar]
- Rokop ME, Auchtung JM, Grossman AD. 2004. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol Microbiol 52: 1757–1767. [DOI] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. 1994. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J 13: 4938–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. 2011. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol 80: 612–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppa J, Kobayashi K, Noirot-Gros MF, Oesterhelt D, Ehrlich SD, Dervyn E, Ogasawara N, Moriya S. 2002. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol Microbiol 45: 59–71. [DOI] [PubMed] [Google Scholar]
- Stray JE, Crisona NJ, Belotserkovskii BP, Lindsley JE, Cozzarelli NR. 2005. The Saccharomyces cerevisiae Smc2/4 condensin compacts DNA into (+) chiral structures without net supercoiling. J Biol Chem 280: 34723–34734. [DOI] [PubMed] [Google Scholar]
- Strick TR, Kawaguchi T, Hirano T. 2004. Real-time detection of single-molecule DNA compaction by condensin I. Curr Biol 14: 874–880. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Hogan E, Koshland D. 1995. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev 9: 587–599. [DOI] [PubMed] [Google Scholar]
- Sullivan NL, Marquis KA, Rudner DZ. 2009. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137: 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JA, Pastrana CL, Butterer A, Pernstich C, Gwynn EJ, Sobott F, Moreno-Herrero F, Dillingham MS. 2015. Specific and non-specific interactions of ParB with DNA: implications for chromosome segregation. Nucleic Acids Res 43: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thadani R, Uhlmann F, Heeger S. 2012. Condensin, chromatin crossbarring and chromosome condensation. Curr Biol 22: R1012–R1021. [DOI] [PubMed] [Google Scholar]
- Umbarger MA, Toro E, Wright MA, Porreca GJ, Bau D, Hong SH, Fero MJ, Zhu LJ, Marti-Renom MA, McAdams HH, et al. 2011. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell 44: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JK, Marquis KA, Rudner DZ. 2009. SirA enforces diploidy by inhibiting the replication initiator DnaA during spore formation in Bacillus subtilis. Mol Microbiol 73: 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Montero Llopis P, Rudner DZ. 2014a. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc Natl Acad Sci 111: 12877–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tang OW, Riley EP, Rudner DZ. 2014b. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Curr Biol 24: 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm L, Bürmann F, Minnen A, Shin H-C, Toseland CP, Oh B-H, Gruber S. 2015. SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. eLife 4: e06659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Errington J. 1994. Bacillus subtilis spoIIIE protein required for DNA segregation during asymmetric cell division. Science 264: 572–575. [DOI] [PubMed] [Google Scholar]
- Youngman PJ, Perkins JB, Losick R. 1983. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci 80: 2305–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.