Abstract
Many cellular RNAs require modification of specific residues for their biogenesis, structure, and function. 5-methylcytosine (m5C) is a common chemical modification in DNA and RNA but in contrast to the DNA modifying enzymes, only little is known about the methyltransferases that establish m5C modifications in RNA. The putative RNA methyltransferase NSUN6 belongs to the family of Nol1/Nop2/SUN domain (NSUN) proteins, but so far its cellular function has remained unknown. To reveal the target spectrum of human NSUN6, we applied UV crosslinking and analysis of cDNA (CRAC) as well as chemical crosslinking with 5-azacytidine. We found that human NSUN6 is associated with tRNAs and acts as a tRNA methyltransferase. Furthermore, we uncovered tRNACys and tRNAThr as RNA substrates of NSUN6 and identified the cytosine C72 at the 3′ end of the tRNA acceptor stem as the target nucleoside. Interestingly, target recognition in vitro depends on the presence of the 3′-CCA tail. Together with the finding that NSUN6 localizes to the cytoplasm and largely colocalizes with marker proteins for the Golgi apparatus and pericentriolar matrix, our data suggest that NSUN6 modifies tRNAs in a late step in their biogenesis.
Keywords: NSUN protein, methyltransferase, 5-methylcytosine, tRNA, RNA modification
INTRODUCTION
Chemical modifications of ribonucleosides play pivotal roles in the maturation, structure, and function of many cellular RNAs. The most heavily modified types of RNA are transfer (tRNAs) and ribosomal RNAs (rRNAs). The majority of rRNA modifications are introduced cotranscriptionally by small nucleolar RNA–protein particles (snoRNPs) (Watkins and Bohnsack 2012), while modifications in tRNAs occur post-transcriptionally and are mainly mediated by lone-standing enzymes (Motorin and Helm 2011; Hopper 2013; Hori 2014). tRNA genes are transcribed by polymerase III as precursor molecules (pre-tRNA) that undergo end processing and numerous modification steps to mature. Nuclear tRNA processing starts with removal of the 5′-leader sequence by RNase P and trimming of the 3′-trailer sequence (Phizicky and Hopper 2010; Betat et al. 2014). In eukaryotes, 3′ end tRNA processing occurs predominantly endonucleolytically and is best studied in Saccharomyces cerevisiae, where these cleavage reactions are carried out by the endonuclease tRNAse Z (Trz1) (Chen et al. 2005; Skowronek et al. 2014), while in humans, the two tRNAse Z homologs, ELAC1 and ELAC2, harbor tRNAse Z activity in vitro (Takaku et al. 2003). ELAC2 was suggested to be involved in nuclear and mitochondrial tRNA 3′ end processing in vivo, whereas the cellular role of ELAC1 remains unclear (Rossmanith 2011). A subset of pre-tRNAs contains introns that are removed during nuclear tRNA splicing reactions in mammals (Paushkin et al. 2004). Prior to export to the cytoplasm, the nucleotidyltransferase Trnt1 adds a nontemplated CCA trinucleotide to the 3′ end of the tRNA that acts as a prerequisite for aminoacylation (Reichert et al. 2001). Although a few aminoacyl-tRNA-synthetases are present in human nuclei (Nathanson and Deutscher 2000), charging of tRNAs with amino acids by aminoacyl-tRNA-synthetases mainly takes place after export to the cytoplasm, where the mature tRNAs act in translation.
Post-transcriptional modification of tRNAs occurs at several steps during their biogenesis, however, the majority of the known mammalian tRNA modification enzymes are found in the nucleus. tRNAs harbor the highest diversity of modifications, of which the majority are methylations or derivatives of methylated residues. 5-methylcytosine (m5C) is a well-characterized modification in DNA. It is also found in RNAs, but only little is known about enzymes that introduce these RNA modifications. The human genome encodes for seven putative m5C RNA methyltransferases (MTases) of the Nol1/Nop2/SUN domain (NSUN) family, which contain the characteristic SUN domain. Introduction of m5C methylations by these enzymes involves formation of a covalent intermediate between a conserved cysteine and the RNA (King and Redman 2002), but only few RNA targets of these enzymes have been identified so far.
NSUN2 is the best characterized m5C RNA MTase to date and it is one of two known m5C RNA methyltransferases that possesses target specificity for tRNAs. The second m5C tRNA methyltransferase described so far, is the DNA methyltransferase 2 (DNMT2), a highly conserved enzyme that structurally resembles DNA methyltransferases, but a role for DNMT2 as a DNA MTase is controversially discussed (Phalke et al. 2009; Schaefer and Lyko 2010; Raddatz et al. 2013). However, DNMT2 was shown to catalyze m5C formation in RNA and the cytosine at position 38 in the anticodon loop of tRNAAsp was identified as a major target (Goll et al. 2006), although in some organisms additional tRNAs were identified as minor substrates (Schaefer et al. 2010; Becker et al. 2012; Tuorto et al. 2012; Mueller et al. 2013; Shanmugam et al. 2014).
In contrast, bisulfite analysis has revealed a broad target spectrum for NSUN2, which can methylate various tRNAs in positions 34, 48, 49, and 50 (Blanco et al. 2014). In addition, NSUN2 methylation targets in vault RNAs and mRNAs have recently also been proposed (Hussain et al. 2013a). It was further shown that the combined loss of both NSUN2 and DNMT2 leads to reduction of general protein synthesis in mice (Tuorto et al. 2012), and that the absence of either of the two proteins reduces tRNA stability (Schaefer et al. 2010; Blanco et al. 2014). Mechanistically, it is suggested that aberrant accumulation of fragmented tRNA upon loss of NSUN2 in patients and mice contribute to neurological disease development (Blanco et al. 2014). In addition, expression of NSUN2 was found to be up-regulated in different tumors (Frye and Watt 2006; Hussain et al. 2009, 2013b).
Similarly, deletions or mutations of NSUN1/p120, NSUN3, NSUN5/WBSCR20, and NSUN7 are also connected to human diseases such as cancer (Freeman et al. 1991; Fonagy et al. 1994, 1995; Bocker et al. 1995; Blagosklonny et al. 1998; Botticelli et al. 1998; Job et al. 2010), developmental disorders (Doll and Grzeschik 2001; Merla et al. 2002; Schubert 2009), or male sterility (Harris et al. 2007; Khosronezhad et al. 2014). Although the target spectrum of these family members is less well studied, the yeast homologs of NSUN1 and NSUN5, Nop2 and Rcm1, have been shown to methylate residues in the 28S ribosomal RNA at positions C2870 and C2278, respectively (Sharma et al. 2013). In humans, knockdown of NSUN1 causes severe pre-rRNA processing defects, whereas depletion of NSUN5 leads to only mild defects in the biogenesis of the large ribosomal subunit (Sloan et al. 2013; Tafforeau et al. 2013; Schosserer et al. 2015). Another member of the NSUN family, NSUN4 was recently found to localize to mitochondria and to methylate C911 in the mitochondrial 12S rRNA in mice (Metodiev et al. 2014). NSUN4 interacts with the mitochondrial transcription termination factor MTERF4 and this interaction is essential for mitochondrial ribosome assembly (Cámara et al. 2011; Metodiev et al. 2014).
For NSUN6, however, its molecular targets and function have remained unknown. Here we have used UV crosslinking and analysis of cDNA (CRAC) and identified tRNACys and tRNAThr as direct interaction partners of NSUN6 in vivo. We found that NSUN6 mediates the methylation of C72 close to the 3′ end of the acceptor stem of tRNACys and tRNAThr. Interestingly, introduction of the modification by NSUN6 requires the presence of the 3′-CCA on its tRNA targets. Together with our finding that NSUN6 localizes to the cytoplasm, these data suggest that NSUN6 modifies tRNAs in a late step of their biogenesis.
RESULTS AND DISCUSSION
Human NSUN6 crosslinks to a subset of tRNAs
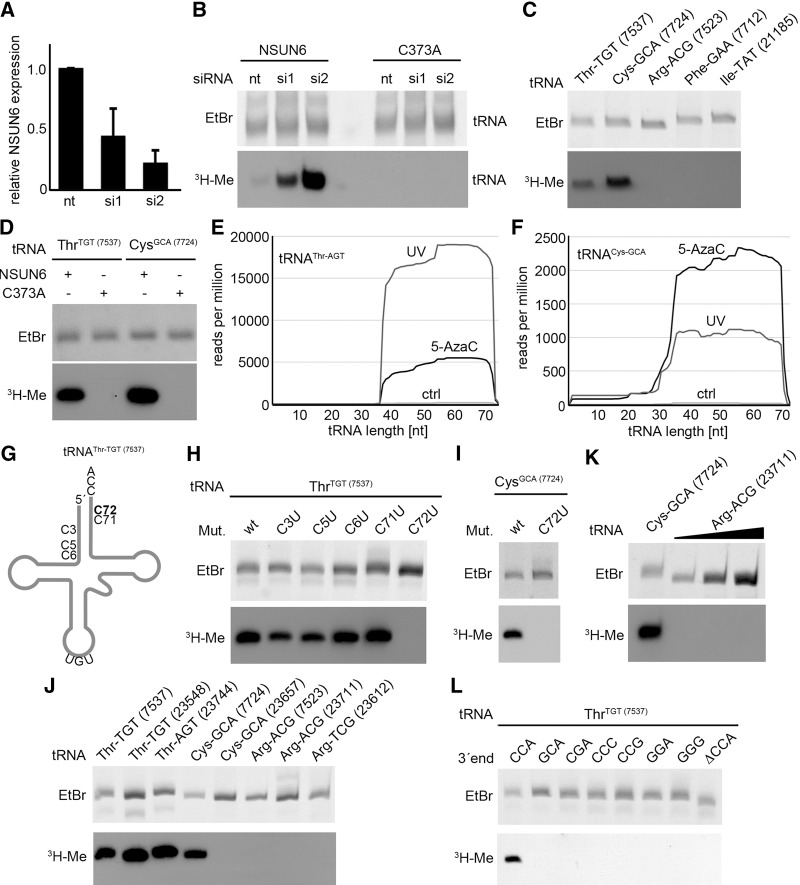
Based on its sequence and predicted domain structure, with an N-terminal pseudouridine synthase and archaeosine transglycosylase (PUA) RNA binding domain and a C-terminal S-adenosyl methionine (SAM) binding domain (Fig. 1A), the NSUN6 protein was suggested to be an m5C RNA methyltransferase (MTase). However, the cellular function of NSUN6 has remained uncharacterized so far. To reveal the molecular target spectrum of NSUN6, we first investigated whether the protein interacts with RNAs in vivo. We therefore generated a stable HEK293 cell line that inducibly expresses NSUN6 with a C-terminal His-PreScission protease site-FLAG (HisPrcFLAG)-tag and performed UV crosslinking followed by purification of complexes and subsequent 32P end labeling of the bound RNA. In comparison to control cells expressing only the HisPrcFLAG-tag, a strong signal indicating copurification of RNA was detected in the samples containing NSUN6 fusion proteins (Fig. 1B). In addition, we performed these experiments using 5-azacytidine (5-AzaC) as a chemical crosslinking reagent (Khoddami and Cairns 2013). 5-AzaC is incorporated into RNA as a cytidine substituent during transcription and specifically traps m5C MTases on their RNA targets in the covalent intermediate during the methylation reaction. Treatment of cells with 5-AzaC also resulted in a strong crosslinking signal with the NSUN6 protein (Fig. 1B), indicating that the covalent intermediate is formed between NSUN6 and cellular RNA. To confirm the specificity of our crosslinking reaction, we next generated a catalytically inactive NSUN6 mutant by replacement of the catalytic cysteine in the TCT tripeptide in motif VI by alanine (C373A) (Fig. 1A). Repetition of the original UV-crosslinking experiment resulted in a signal for radiolabeled RNA crosslinked to the NSUN6 C373A mutant, indicating that the protein is folded correctly and that the mutation does not impair target RNA binding (Fig. 1B). In contrast, no radioactive signal could be detected after chemical crosslinking with 5-AzaC, demonstrating that the NSUN6-C373A mutant cannot form the covalent protein–RNA intermediate and thus confirming that the mutant is catalytically inactive (Fig. 1B).
FIGURE 1.
NSUN6 crosslinks to tRNAs in vivo. (A) Schematic view of human NSUN6. The black line represents the NSUN6 protein; the predicted PUA RNA binding domain and the methyltransferase (MTase) domain are drawn as black boxes. The magnified view shows the amino acid sequence of motif VI in the MTase domain. The C373 (indicated as bold capital letter) was mutated to generate the catalytically inactive mutant. (B) Cells expressing NSUN6-HisPrcFLAG (NSUN6), the catalytically inactive mutant NSUN6-C373A-HisPrcFLAG (C373A), or only the tag (FLAG) as a control were treated with 5-azacytidine (5-AzaC) or crosslinked using 254 nm light (UV) as indicated. The tagged proteins and crosslinked RNAs were purified and the RNA was radioactively labeled, followed by separation of RNA–protein complexes by SDS-PAGE, transfer to a nitrocellulose membrane, and exposure to an X-ray film. (C) UV or 5-AzaC crosslinking and analysis of cDNA (CRAC) experiments were performed for sequence library production and Illumina deep sequencing. The exposure of an X-ray film after complex purification, RNA linker ligations, radiolabeling, and SDS-PAGE of the RNA–protein complexes is shown. The black boxes indicate the parts of the membrane containing the radiolabeled RNA that were cut out and from which the RNA was extracted and processed for Illumina deep sequencing. (D–F) Pie charts present different RNA classes and the relative distribution of Illumina sequence reads, which were obtained after UV crosslinking of cells expressing either the HisPrcFLAG tag (D) or NSUN6-HisPrcFLAG (E) or following 5-AzaC crosslinking of the NSUN6-HisPrcFLAG cell line (F). (tRNA) transfer RNA, (snRNA) small nuclear RNA, (snoRNA) small nucleolar RNA, (rRNA) ribosomal RNA, (mtRNA) mitochondrial encoded RNA, (miscRNA) miscellaneous RNA, (miRNA) microRNA, (lncRNA) long noncoding RNA.
To identify the target RNAs of the human NSUN6 protein, we performed UV Crosslinking and Analysis of cDNA (CRAC). After UV crosslinking in vivo, protein–RNA complexes were purified; the bound RNA trimmed, radiolabeled, and ligated to adapters before SDS-PAGE and transfer to a membrane (Fig. 1C). The RNA extracted from the membrane was used for RT-PCR to generate a sequence library for Illumina deep sequencing. Obtained sequences were mapped on the human genome and the comparison of the results with the control revealed that tRNAs were strongly overrepresented in the data from the NSUN6 sample. Here, >40% of all sequence reads were mapped to sequences encoding tRNAs, indicating that NSUN6 predominantly interacts with tRNAs in vivo (Fig. 1D,E). We then performed the CRAC experiment using 5-AzaC as crosslinking reagent (5-AzaC CRAC) and the data obtained confirmed the results of the UV CRAC experiment (Fig. 1F). Together these results suggest that NSUN6 is an m5C tRNA methyltransferase.
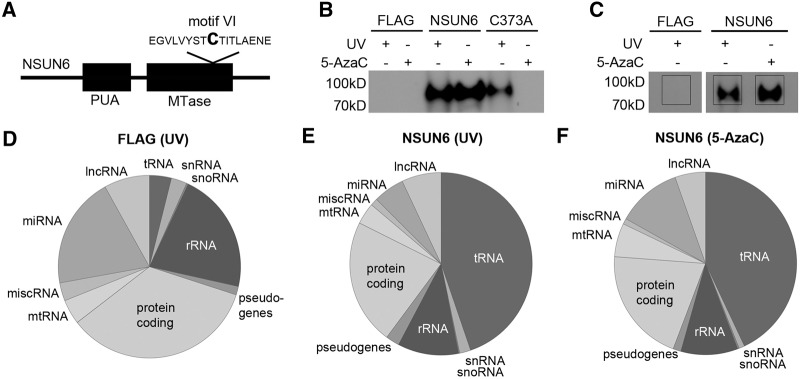
Detailed analysis of sequences mapped to tRNAs revealed the specific enrichment of tRNACys, tRNAThr, and tRNAArg in the UV CRAC experiment, while the HisPrcFLAG control showed an equal distribution of tRNAs (Fig. 2A). Interestingly, both tRNACys and tRNAThr were equally enriched after 5-AzaC CRAC, however, fewer sequences mapping to tRNAArg were found than in the UV CRAC (Fig. 2A; Supplemental Material). To confirm the interaction between NSUN6 and these tRNA species, we performed UV crosslinking followed by pulldown of the NSUN6-HisPrcFLAG and HisPrcFLAG control proteins. Bound RNA was extracted from the complexes, separated on a polyacrylamide gel and analyzed by Northern blot using probes against the tRNA candidates and control tRNAs that were not enriched in the CRAC experiments. Since the mapping of the CRAC sequences resulted in peaks on several isoacceptors and isodecoders of the tRNAs we generated our probes against one representative isoacceptor of each tRNA type that show substantial enrichment with the NSUN6 samples. This confirmed the interaction of NSUN6 with tRNACys-GCA (7724) and tRNAThr-TGT (7537) (Fig. 2B, upper panels). Only after prolonged exposure a very weak signal for tRNAArg-ACG (7523) could be detected. Probes against tRNAPhe-GAA (7712) and tRNAIle-TAT (21185), which were not enriched in the CRAC experiment (see Supplemental Material), were used as controls (Fig. 2B, lower panels) and no signal was observed for these tRNAs even after prolonged exposure. We conclude that NSUN6 interacts with full-length tRNACys and tRNAThr in vivo and only weakly with tRNAArg.
FIGURE 2.
NSUN6 binds to a specific subset of tRNAs. (A) The relative abundance of sequence reads from different tRNAs obtained in CRAC experiments using UV or 5-AzaC crosslinking with cells expressing the NSUN6-HisPrcFLAG (NSUN6) protein or control cells (FLAG) is shown graphically. The percentage of hits for all isoacceptors of the listed tRNA species is given. The asterisk (*) marks tRNAs that are not enriched in the NSUN6 samples. This includes, from bottom to top, tRNAMet, Leu, Gly, Glu, Ser, Asp, Lys, Ala, Pro, Val, tRNA pseudogenes, and tRNAIle, Gln, Asn, Tyr, His, Trp, Phe; the hash (#) indicates tRNAHis, Trp, Phe. (B) UV crosslinking and pulldown of the NSUN6 associated RNA was performed as described in Figure 1. The RNA was precipitated from the isolated protein–RNA complexes, separated on a denaturing polyacrylamide gel and transferred to a nylon membrane. Different tRNA probes against the indicated isoacceptors identified in the CRAC experiments were used for Northern blot analysis. Input: 1%, elution: 50%.
NSUN6 methylates tRNAThr and tRNACys
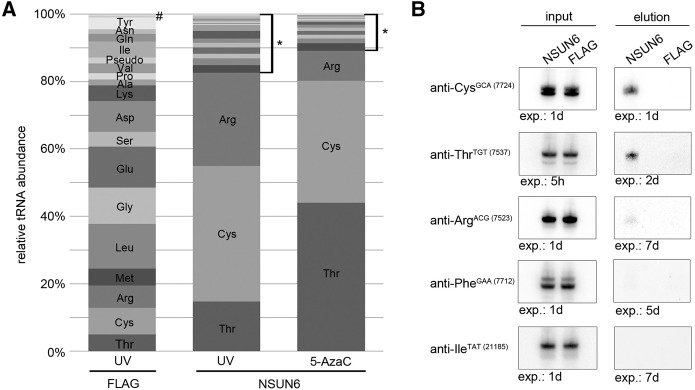
To gain further insight into the catalytic specificity of NSUN6, we investigated its methylation activity in vitro. Recombinant His14-MBP-NSUN6 and the corresponding NSUN6-C373A catalytically inactive mutant were expressed and purified from Escherichia coli. For in vitro methylation assays, total RNA from cells treated with nontarget siRNAs or those targeting NSUN6 (Fig. 3A) was incubated with recombinant NSUN6 in the presence of 3H-labeled S-adenosyl methionine (3H-SAM) and the RNA was subsequently separated on a polyacrylamide gel. Methylated substrates could be identified after exposure of the dried gel to an X-ray film. RNA in the size range of tRNA that was isolated from the NSUN6 knockdown cells could be methylated by wild-type NSUN6 in vitro, while only very weak methylation of RNA extracted from cells treated with nontarget siRNA was observed (Fig. 3B). However, incubation with the catalytically inactive mutant of NSUN6 (C373A) did not lead to methylation of the extracted tRNAs from any of the cell lines, demonstrating that the mutant is indeed inactive and that no active RNA methyltransferase is copurified with NSUN6 from E. coli. These data indicate that knockdown of NSUN6 leads to a reduction in the methylation state of NSUN6 target RNAs in vivo, which then allows incorporation of 3H-labeled methyl groups upon treatment with recombinant NSUN6 in vitro. In contrast, RNA from nontarget siRNA treated cells could not be methylated in vitro, suggesting that the target RNAs already contain the corresponding methylations. These data confirm that NSUN6 is an active RNA methyltransferase in vivo and in vitro.
FIGURE 3.
NSUN6 methylates tRNAThr and tRNACys at position C72 in vitro. (A) HeLa cells were transfected with two different siRNAs (si1, si2) against NSUN6 or with nontarget (nt) siRNA. The knockdown efficiency was analyzed by quantitative PCR. The relative abundance of the NSUN6 mRNA was normalized to the mean of GAPDH and DDX21 mRNA levels. Data are presented as mean ± SD. (B) Total RNA isolated from NSUN6 knockdown (si1 or si2) or control cells (nt) was used for in vitro methylation assays with recombinant His14-MBP-NSUN6 or the catalytically inactive mutant His14-MBP-NSUN6-C373A and tritiated S-adenosyl-methionine as the methyl group donor. RNA was separated by denaturing PAGE before transfer onto a nylon membrane and autoradiography. Exposure time: 14 d. (C) Putative tRNA substrates identified in the CRAC experiments and control tRNAs not enriched with NSUN6 were analyzed using in vitro methylation experiments with in vitro transcribed tRNAs and recombinant NSUN6. Exposure time: 1 d. (D) In vitro methylation reactions of tRNAThr-TGT (7537) and tRNACys-GCA (7724) were performed using recombinant NSUN6 wild-type protein or the NSUN6-C373A mutant. Exposure time: 5 d. (E,F) The mapping of NSUN6 associated CRAC sequence reads on the tRNA substrates tRNAThr-AGT (E) and tRNACys-GCA (F) is shown. Reads from the NSUN6-HisPrcFLAG are presented with UV-crosslinking (UV) in dark gray and with 5-azacytidine (5-AzaC) crosslinking in black. The control (ctrl) sample (HisPrcFLAG) is shown in light gray. (G) Schematic presentation of a tRNAThr-TGT with labeled acceptor stem cytosine residues, which were mutated for NSUN6 target residue identification. The nucleoside modified by NSUN6, C72, is shown in bold. (H) Cytosine to uracil point mutations were introduced separately for all cytosines in the acceptor stem of the tRNAThr-TGT (7537), and the corresponding tRNA transcripts were analyzed by in vitro methylation assays with recombinant NSUN6. Exposure time: 1 d. (I) In vitro methylation analysis of the tRNACys-GCA (7724) wild-type tRNA and the corresponding C72U mutant was performed with NSUN6. Exposure time: 1 d. (J) In vitro methylation analysis was carried out using NSUN6 and different isoacceptors of tRNACys, tRNAThr, and tRNAArg. The tRNACys-GCA (7724) isoacceptor occurs naturally without a C72. (K) Synthetic tRNACys-GCA (7724) and increasing amounts of tRNAArg-ACG (23711) were subjected to in vitro methylation by NSUN6. Chemically synthesized RNA was used to avoid using in vitro transcripts that might contain nontemplated nucleotides introduced by T7 polymerase. (L) In vitro methylation analysis of CCA mutants of tRNAThr-TGT (7537). The CCA tail was either deleted, or one, two, or all three of the nucleotides of the CCA were exchanged. Exposure time: 1 d.
To identify the individual substrates of NSUN6, we generated transcripts of tRNACys, tRNAThr, tRNAArg, tRNAIle, and tRNAPhe in vitro and subjected them to methylation assays with recombinant NSUN6. tRNAThr and tRNACys showed strong methylation, indicating that these two tRNAs are major NSUN6 substrates, while no methylation signal was detected for tRNAPhe and tRNAIle (Fig. 3C). For tRNAArg, no methylation could be detected even after prolonged exposure, implying that tRNAArg is not an NSUN6 substrate. Moreover, this in vitro assay also revealed that NSUN6 can methylate its substrate tRNAs, tRNAThr, and tRNACys, without any previous modifications and that the RNA methyltransferase does not depend on interactions with other proteins or cofactors for its activity. The specificity of the methylation reaction on the in vitro transcribed tRNAThr and tRNACys was further confirmed using the C373A mutant, resulting in no methylation signal after incubation of the tRNAs with the mutant protein (Fig. 3D). Together with the observation that mutation of NSUN6 at C373A prevents formation of the covalent intermediate in 5-AzaC crosslinking experiments (Fig. 1B), the lack of in vitro methylation by NSUN6 C373A strongly suggests that NSUN6 functions using the characteristic m5C RNA methyltransferase-like mechanism that relies on the cysteine residue of the TCT in motif VI. In contrast, DNA methyltransferases utilize a different mechanism involving a cytosine in the conserved PCG peptide in motif IV for covalent complex formation during the methylation reaction (Santi et al. 1984; Wu and Santi 1987; King and Redman 2002). Remarkably, m5C methylations in tRNA can be mediated by both mechanisms, since DNMT2 resembles the structure of a DNA methyltransferase and also uses the DNA methylation mechanism for tRNA modification (Jurkowski et al. 2008).
Methylation of tRNAThr and tRNACys by NSUN6 occurs at position C72
In order to identify the target residue for NSUN6 we analyzed the peak distribution in our CRAC data for the tRNAThr and tRNACys genes. Here, we found that the reads obtained from the NSUN6 samples predominantly map to the 3′ ends of the tRNAs (Fig. 3E,F). We therefore performed site-directed mutagenesis on tRNAThr-TGT (7537) generating cytosine to uracil mutants of all cytidines in the acceptor stem. Interestingly, all the tRNA mutants except for the C72U mutant could be methylated in in vitro assays, indicating that the C72 is the target nucleotide (Fig. 3G,H). By similar mutational analysis, it could be confirmed that C72 is also the targeted residue in the tRNACys-GCA (7724) substrate (Fig. 3I).
Since the CRAC data identified several isoacceptors of the substrate tRNAs, we investigated the substrate specificity of the candidate tRNA species in further detail. For tRNAThr we cloned an additional isoacceptor with TGT anticodon that was only slightly enriched with NSUN6 and one possessing a different anticodon (AGT). Methylation assays with the corresponding in vitro transcripts showed that for tRNAThr all analyzed isoacceptors could be methylated with similar efficiency (Fig. 3J). tRNACys only exists with the GCA codon and most of these tRNAs have similar sequences. However, few tRNACys-GCA isoacceptors naturally occur without a C72. We therefore included such an isoacceptor in our analysis and found that this tRNA was not methylated by NSUN6 (Fig. 3J), providing further confirmation that the cytosine 72 is the target nucleotide of the methyltransferase. In the case of the tRNAArg-ACG, we focused our analysis on the two isodecoders with strongest enrichment in the CRAC experiment as well as one prominent candidate with TCG anticodon. All isoacceptors tested contain a C72 but none of them was methylated (Fig. 3J), indicating that tRNAArg-GCG is not a methylation substrate of NSUN6. This was further confirmed by in vitro methyltransferase assays using synthetic tRNAArg-ACG (23711), and tRNACys-GCA (7724) as a positive control. Here, in vitro methylation of these synthetic tRNAs resulted in a strong methylation signal for tRNACys, but no methylation could be observed even with increasing amounts of the synthetic tRNAArg (Fig. 3K). While misfolding of nonmodified tRNAArg cannot be excluded, these data indicate that the lack of methylation of tRNAArg by NSUN6 is not due to incorrect termination and addition of nontemplated nucleotides by the T7 polymerase at the 3′ end of the tRNA. Taken together, the observations that the tRNAArg isoacceptors, the tRNAPhe-GAA (7712) and tRNAIle-TAT (21185) transcripts that were included in our analysis, also contain a C72 and show no methylation by NSUN6 (Fig. 3C,J,K); these data suggest that the presence of a C72 alone is not sufficient to specify a tRNA as an NSUN6 substrate.
The mapping of m5C modifications across the human transcriptome by bisulfite sequencing previously revealed the m5C methylation status of several tRNAs (Squires et al. 2012). For example, tRNAThr-TGT showed conversion rates of the C72 nucleotide of <30%, demonstrating that this nucleotide is methylated in vivo. This is in line with the results from in vitro methylation assays using total cellular RNA (Fig. 3B), where methylation by NSUN6 is only observed for RNA isolated from cells that were depleted of NSUN6, suggesting that its RNA targets are normally methylated in vivo to a high extent. In contrast to the bisulfite sequencing of tRNAThr-TGT, none of the analyzed tRNAArg isoacceptors showed conversion of <80% at the position C72, suggesting that this position is unmodified (Squires et al. 2012). Similar results were obtained for tRNAMet-CAT and tRNAGlu-TTC, two additional tRNAs that contain a C72. These data support our finding that only a subset of the C72 containing tRNAs is targeted by NSUN6. Similarly, DNMT2 and NSUN2 do not methylate every tRNA containing their target nucleotides. Only very few tRNAs that contain a C38 are substrates of DNMT2 and tRNAAsp-GTC is with one exception the major substrate in all investigated organisms (Goll et al. 2006; Schaefer et al. 2010; Tovy et al. 2010; Becker et al. 2012; Mueller et al. 2013; Shanmugam et al. 2014). Additional tRNAs like tRNAGlu, tRNAVal, and tRNAGly were found to represent targets only in some species (Schaefer et al. 2010; Becker et al. 2012; Tuorto et al. 2012; Mueller et al. 2013). Although DNMT2 and NSUN6 seem to be multisubstrate RNA MTases they exhibit a narrow target spectrum. In contrast, NSUN2 has a broad target spectrum in mice and humans (Blanco et al. 2014). Surprisingly, the NSUN6 target tRNACys does not appear to be a substrate of NSUN2 although almost all isoacceptors contain C48, 49, and 50. We conclude that NSUN6 is a multisubstrate m5C RNA MTase that methylates position C72 in a specific subset of tRNAs.
Methylations mediated by NSUN6 require the presence of a 3′-CCA in the tRNA substrates
Upon closer inspection of our CRAC data, we observed that nearly all reads covering the 3′ end of NSUN6 bound tRNAs contain the terminal CCA. To investigate the importance of a 3′-CCA for target recognition, we generated a tRNAThr-TGT (7537) mutant lacking the CCA at the 3′ end. Strikingly, the CCA deletion mutant could not be methylated by NSUN6, suggesting that the CCA is important for target recognition (Fig. 3L, right-hand lane). We then generated six different tRNAThr-TGT (7537) mutants by exchanging one, two, or all three nucleotides of the CCA-tail. None of these mutants were methylated by NSUN6 (Fig. 3L), suggesting that NSUN6 specifically recognizes the presence of the CCA sequence.
The NSUN6 protein contains a pseudouridine synthase and archaeosine transglycosylase (PUA) domain that is highly conserved and found in a wide range of archaeal, bacterial, and eukaryotic proteins (Fig. 1A; Cerrudo et al. 2014). Proteins that contain PUA domains function in enzymatic RNA modification like pseudouridine synthases or tRNA modifying enzymes (Cerrudo et al. 2014). The crystal structure of the Pyrococcus horikoshii archaeosine tRNA-Guanine transglycosylase (ArcTGT) shows that the PUA domain directly contacts the RNA substrate and contributes to the specificity of the protein (Ishitani et al. 2002). tRNA binding by the ArcTGT involved direct contact to the CCA via a specific surface formed by the PUA domain. Our data show that not only the presence of three nucleotides at the 3′ end of the tRNA but also the specific CCA sequence is required for target recognition by NSUN6. This suggests that the tRNA binding pocket of NSUN6 binds these residues in a sequence-dependent manner. In contrast, target recognition by DNMT2 is independent of the tRNA ends. Methylation of C38 can take place without the CCA tail and even additional nucleotides at the ends of the tRNA do not inhibit enzyme activity in vitro (Mueller et al. 2013). Here, a sequence pattern (C32, A37, C38, C40) was suggested to be required for target recognition by DNMT2 (Mueller et al. 2013) and it was recently shown that a GG dinucleotide in the variable loop works as an anti-determinant for DNMT2 methylation (Shanmugam et al. 2014). The formation of m5C34 in tRNALeu-CAA by NSUN2 requires the presence of the intron in the pre-tRNA (Brzezicha et al. 2006). In the case of NSUN6, the CCA tail contributes to target recognition but it is so far not known, which additional sequence or structural information of the tRNA is required for its recognition as an NSUN6 substrate. Both identified NSUN6 targets, tRNAThr and tRNACys, share a common C71 (Fig. 3G), but according to our mutational analysis this nucleotide is not important for target specification.
NSUN6 localizes in the cytoplasm of human cells
The finding that tRNA methylation by NSUN6 requires a CCA at the 3′ end of its substrates implies that the NSUN6-mediated modification occurs at a late stage in tRNA biogenesis. We therefore analyzed the subcellular distribution of NSUN6 by fractionating HEK293 cells expressing the NSUN6-HisPrcFLAG fusion protein into a nuclear and a cytoplasmic extract. Interestingly, Western blot analysis revealed that the NSUN6 fusion protein is found exclusively in the cytoplasm (Fig. 4A). Antibodies against UTP14A and tubulin were used as nuclear and cytoplasmic controls, respectively. These data indicate that NSUN6 localizes to the cytoplasm, while the majority of all known tRNA modifications are established in the nucleus. In the next step, we generated stable HEK293 cell lines that express tetracycline inducible GFP-NSUN6 or NSUN6-GFP fusion proteins and analyzed their subcellular localization by fluorescence microscopy. Interestingly, both NSUN6 fusion proteins indeed localized to the cytoplasm, where they were found as a weak dispersed cytoplasmic staining, however, specific substructures could also be detected (Fig. 4B,C). By combining the fluorescence microscopy for GFP-tagged NSUN6 with immunofluorescence against marker proteins for pericentriolar matrix (PCM) and microtubule organization center (MTOC), PCM1 and γ-tubulin, as well as the cis-Golgi marker protein GM130 or the trans-Golgi marker GOPC, we could detect colocalization of the cis-Golgi and PCM markers with the structures stained by both GFP-NSUN6 and NSUN6-GFP fusion proteins. The staining pattern of NSUN6 matches the immunostaining obtained with the PCM markers and the GM130 antibody better than the pattern of the GOPC protein (Fig. 4B,C), suggesting that a large fraction of NSUN6 localizes to and around the PCM. Interestingly, NSUN2 was previously shown to bind the mitotic spindle in mitosis, however, it is a nucleolar protein in interphase, indicating that there is likely no link between its recruitment to the mitotic spindle and the localization of NSUN6 at the PCM in interphase (Frye and Watt 2006; Hussain et al. 2009). Besides the nucleolar NSUN2 in interphase, also DNMT2 localizes predominantly to the nucleus or is associated with the nuclear matrix (Banerjee et al. 2005; Kuhlmann et al. 2005; Schaefer et al. 2008). Only ectopically expressed DNMT2 with N-terminal myc-tag showed cytoplasmic localization (Goll et al. 2006). The localization of NSUN6 in the cytoplasm and its partial recruitment to the Golgi apparatus was unexpected as, to our knowledge, this is the first report indicating the presence of an RNA methyltransferase at the PCM and Golgi in interphase. While the role of its recruitment to Golgi membranes remains unclear it is possible that tethering of a fraction of the RNA methyltransferase to structures in the cytoplasm might play a role in keeping NSUN6 cytoplasmic. Reduction of the free pool of proteins that could otherwise enter the nucleus by diffusion through nuclear pore complexes or while the nuclear envelope is reassembled at the end of mitosis, has previously been described to play an important role in retaining certain proteins in the cytoplasm (see, for example, Wang et al. 2000; Ohtsubo et al. 2009). It might also be important that NSUN6 is excluded from nuclei so that it only encounters its substrate tRNAs after their nuclear maturation and export to the cytoplasm.
FIGURE 4.
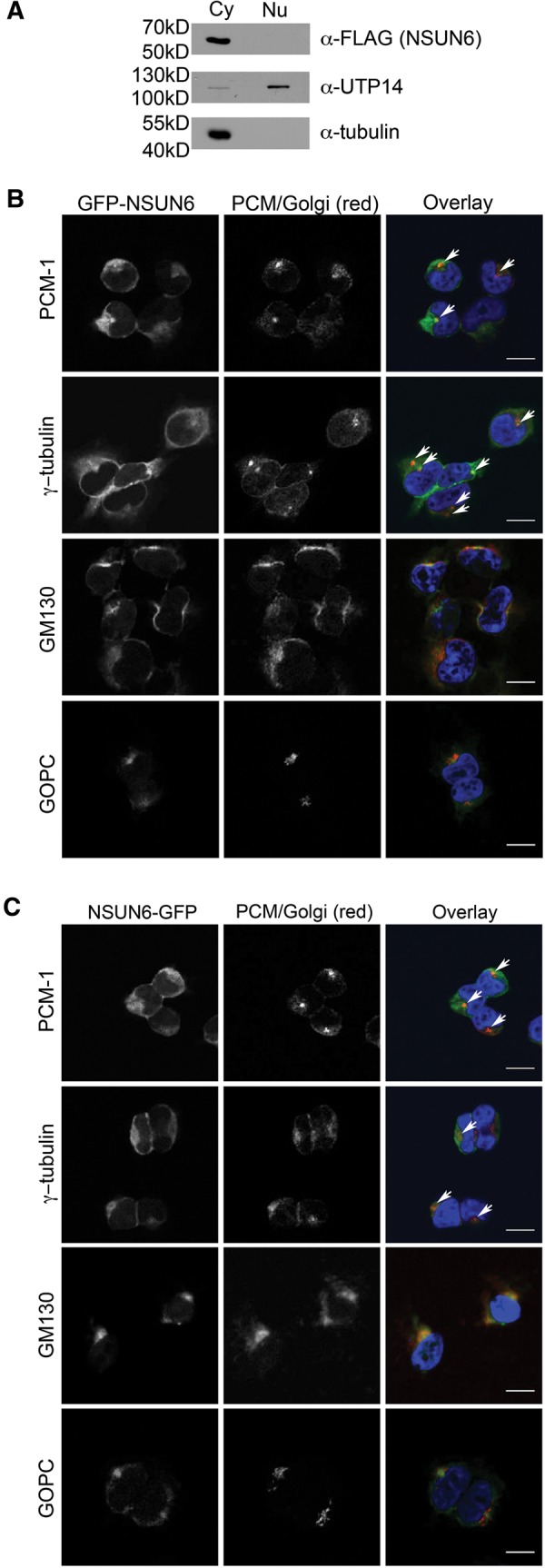
NSUN6 localizes to the cytoplasm and partially colocalizes with pericentriolar matrix and Golgi apparatus marker proteins. (A) HEK293 cells stably expressing NSUN6-HisPrcFLAG were separated into nuclear and cytoplasmic fractions. Nuclear and cytoplasmic protein extracts from equal numbers of cells were separated by SDS-PAGE and analyzed by Western blot. NSUN6-HisPrcFLAG was detected using an anti-FLAG antibody; UTP14A and tubulin were used as nuclear and cytoplasmic markers, respectively. (B,C) HEK293 cells stably expressing GFP-NSUN6 (B) or NSUN6-GFP (C) under the control of a tetracycline inducible promoter were examined by fluorescence microscopy (green in overlay). The pericentriolar matrix (PCM) or Golgi apparatus (red in overlay) was stained by immunofluorescence using anti-γ-tubulin (MTOC), anti-PCM1 (PCM), anti-GM130 (cis-Golgi), or anti-GOPC (trans-Golgi) antibodies. White arrows indicate colocalization of NSUN6 with γ-tubulin (MTOC) and PCM1 (PCM). Scale bar: 20 µm.
In summary, we have identified NSUN6 as a novel mammalian m5C tRNA methyltransferase. Furthermore, our data suggest that methylation of tRNAs by NSUN6 occurs during a late step of tRNA biogenesis, after the addition of the CCA-tail and export of the tRNA from the nucleus. Target identification of the hitherto uncharacterized NSUN6 protein expands our knowledge on m5C RNA methyltransferases and contributes to a comprehensive view on tRNA methylation. Surprisingly, all three identified m5C tRNA MTases, NSUN2, DNMT2, and NSUN6, have multiple substrates, but they seem to have a differential target spectrum and different localizations. It will be interesting to gain further insight into the distinct and overlapping functions of the three tRNA methyltransferases and whether their modifications are required for efficient aminoacylation of their tRNA targets.
MATERIALS AND METHODS
Human cell culture and generation of stable cell lines
HEK293 Flp-In T-Rex and HeLa CCL2 cells were cultured at 37°C with 5% CO2 in 1× Dulbecco's modified eagle medium (DMEM) supplemented with 10% FCS and 2 mM glutamine. For generation of tetracycline-inducible stable cell lines, the NSUN6 CDS (NM_182543.3) was cloned into the pcDNA5 vector with C-terminal His-PreScission protease cleavage site-2×FLAG tag (HisPrcFlag) or N- or C-terminal GFP tag and transfected into HEK293 Flp-In T-Rex cells (Life Technologies) according to the manufacturer's instructions. To generate the NSUN6 C373A mutant, two point mutations (tgc → gcc) were introduced in the relevant codon by site-directed mutagenesis.
siRNA treatment, RNA isolation and qRT-PCR
HeLa CCL2 cells were transfected with siRNAs (nontarget: 5′-UCGUAAGUAAGCGCAACCC-3′; NSUN6_1 5′-UUAGUAACUGAAUCCGUGGUG-3′ or NSUN6_2 5′-UUCUGGGUCCAAUAACAGGAA-3′) using Lipofectamine RNAiMax (Life Technologies) according to the manufacturer's instructions. Cells were harvested 72 h after siRNA transfection and total RNA was isolated using TRIzol (Life Technologies). The knockdown efficiency was determined by qRT-PCR and relative quantification was performed using GAPDH and DDX21 as housekeeping genes. The primer sequences were: NSUN6_fw 5′-CCAGAGGGTGTGCTGGTTTAT-3′ and NSUN6_rev 5′-TCTCCTCCAATCTGCGGTTC-3′; GAPDH_fw 5′-CTGGCGTCTTCACCACCATGG-3′ and GAPDH_rev 5′-CATCACGCCACAGTTTCCCGG-3′; DDX21_fw 5′-GCAGCAGTTATTGGGGATGTC-3′ and DDX21_rev 5′- AGGGTGATTTCCCTTTGCTTC-3′.
Crosslinking and analysis of cDNA (CRAC)
Crosslinking and analysis of cDNA (CRAC) was carried out as previously described (Bohnsack et al. 2012; Sloan et al. 2015). In brief, HEK293 cells expressing NSUN6-HisPrcFlag, NSUN6-C373A-HisPrcFLAG or the HisPrcFlag tag alone were induced using 1 μg/mL tetracycline for 24 h and UV crosslinking was carried out using a Stratalinker (Stratagene). For chemical crosslinking, cells were incubated in growth media complemented with 5-azacytidine (Sigma), harvested in buffer containing 50 mM Tris/HCl pH 7.6, 150 mM NaCl, 0.1% NP-40, 5 mM β-mercaptoethanol and protease inhibitors, and lysed by sonication. Protein–RNA complexes were tandem affinity purified using anti-FLAG magnetic beads (Sigma) followed by elution with PreScission Protease. The RNA was partially digested (1 min with 1 unit RNace-IT [Agilent]) and radiolabeled using T4 PNK (Thermo Scientific) and [γ-32P]ATP (PerkinElmer); complexes were purified using Nickel-NTA (Qiagen) under denaturing conditions (6 M guanidium-HCL) and transferred to a nitrocellulose membrane after separation by NuPAGE gel electrophoresis. Radioactive signals were detected by exposure to an X-ray film. For Illumina deep sequencing, 3′ and 5′ adapters were ligated to the coimmunoprecipitated RNA, the RNA was reverse transcribed using Superscript III Reverse Transcriptase (Sigma) and the cDNA library was amplified using primers that contained randomized sequences of 5 nt to distinguish PCR templates. Prior to mapping, 3′-adapter sequences and bases with a phred quality score <13 (95% base call accuracy) were removed using Flexbar. Reads with equal sequence and barcode were collapsed using python scripts resulting in 379844 reads for NSUN6 (UV), 414570 (5-AzaC), and 231690 for the FLAG control. Bowtie2 was used to map the remaining reads on the human Ensembl genome version GRCh 37.75 with an 18-nt cutoff.
Detection of crosslinked tRNAs by Northern blotting
For specific detection of coimmunoprecipitated tRNA by Northern blot, crosslinking, and immunoprecipitation experiments were performed as previously described (Sloan et al. 2013). RNA from crosslinked complexes was eluted by Proteinase K digestion for 16 h and the precipitated RNA was resuspended in loading dye (95% formamide, 5 mM EDTA, bromophenolblue), separated on a denaturing 12% polyacrylamide (7 M urea) and transferred to a nylon membrane. Selected tRNAs were detected by Northern blotting using specific probes (anti-Thr-TGT: AGGCCCCAGCGAGATTTGAACTCGCGACCCCTGG, anti-Phe-GAA: TAGATCTTCAGTCTAACGCTCTCCCAACTG, anti-Arg-ACG: CAGGAGTCGAACCTGGAATCTTCTGATCCG, anti-Cys-GCA: GGGGGCACCTGGATTTGAACCAGGGACCTC, anti-Ile-TAT: ACCGCGCGCTAACCGATTGCGCCACTGGAGC) that were 5′ end labeled using T4 PNK (Thermo Scientific) and [γ-32P]ATP (6000Ci/mmol, PerkinElmer) and a phosphorimager.
Cloning and recombinant expression of NSUN6 in E. coli
The NSUN6 CDS (NM_182543.3) was cloned into a pQE80 derivative containing an N-terminal His14-MBP-tag (Weis et al. 2014). The NSUN6 C373A was generated by site-directed mutagenesis as described above. The recombinant proteins were expressed in E. coli (DE3) Rosetta pLysS cells. Induction and protein purification were performed as described previously (Mueller et al. 2013). In brief, expression of NSUN6 was induced with 0.5 mM IPTG for 16 h at 18°C. After harvesting, cells were resuspended in lysis buffer (30 mM KPi pH 7.0, 300 mM KCl, 10% (v/v) glycerol, 10 mM imidazole, 0.1 mM dithiothreitol [DTT]) containing protease inhibitors (complete mini, Roche) and lysed by sonication. The lysate was cleared by centrifugation and the supernatant was incubated with NiNTA-Sepharose followed by washing with lysis buffer and elution with a buffer containing 30 mM KPi pH 7.0, 300 mM KCl, 10% glycerol, 200 mM imidazole, and 0.1 mM DTT. For the second purification step the NiNTA-Sepharose eluate was diluted 1:5 in amylose column buffer (20 mM KPi pH 7.0, 200 mM KCl, 10% [v/v] glycerol, 2 mM β-mercaptoethanol) and incubated with amylose beads. Bound proteins were eluted using column buffer complemented with 10 mM maltose and stored in a buffer containing 30 mM KPi pH 7.0, 100 mM KCl, 50% glycerol, 1 mM DTT, and 0.1 mM EDTA.
Cloning and in vitro transcription of human tRNA genes
tRNAs used in this study are annotated according to the tRNAScan-SE 1.21 (Lowe and Eddy 1997; Schattner et al. 2005). The human tRNAThr-TGT (7537) (chr. 14), tRNAThr-TGT (23548) (chr. 5), tRNAThr-AGT (23744) (chr. 6), tRNACys-GCA (7724) (chr. 3), tRNACys-GCA (23657) (chr. 7), tRNAArg-ACG (7523) (chr. 14), tRNAArg-TCG (23612) (chr. 15), tRNAArg-ACG (23711) (chr. 6), tRNAPhe-GAA (7712) (chr. 11) and tRNAIle-TAT (21185) (chr. 19) were cloned into a pQE vector derivative that does not contain an internal T7 promotor. tRNA genes were produced by recursive PCR as described previously (Mueller et al. 2013) using four overlapping oligonucleotides per gene. The CCA tail and a BsaI restriction site were added at the 3′ end of the tRNA gene ensuring that the generated in vitro transcripts end after the CCA. The forward primer contained the T7 promoter, which was attached to the 5′ end of the tRNA gene. Point mutations were introduced by site-directed mutagenesis. For in vitro transcription, 500 ng of BsaI-linearized plasmid were incubated with 1 mM NTPs, T7-RNA polymerase, 1× transcription buffer (Fermentas) and RNasin RNase Inhibitor (Promega) for 1 h at 37°C. After transcription, samples were treated with DNase I for 15 min and purified over a Sephadex G-25 spin column (Roche).
Preparation of synthetic tRNAs
In addition to the in vitro transcripts tRNACys-GCA (7724) (chr. 3) and tRNAArg-ACG (23711) (chr. 6) were also chemically synthetized. RNA oligonucleotides were prepared by solid-phase synthesis using 2′-O-TOM-protected ribonucleotide phosphoramidites, (ChemGenes) chemically phosphorylated on the solid support, deprotected in two steps with methylamine in water/ethanol, followed by 1 M tetrabutylammonium fluoride in tetrahydrofuran, purified by denaturing PAGE, and analyzed by analytical anion exchange chromatography and ESI-MS. Synthetic tRNAs were prepared by enzymatic ligation of chemically synthesized RNA fragments using T4 DNA ligase (Fermentas) and DNA splint oligonucleotides (2–5 nmol scale, incubation at 30°C for 12 h), analogous to previously reported procedures (Rieder et al. 2009). The full-length tRNAs were isolated by denaturing PAGE, extracted into Tris—NaCl buffer, precipitated with ethanol and re-dissolved in water.
3H-in vitro methylation assay
Methylation of RNA substrates was carried out as described previously with minor modifications (Jurkowski et al. 2008; Mueller et al. 2013). Reactions containing 1 µM recombinant NSUN6 and 1 µM of tRNA-transcript (0.3, 1, and 2 µM with synthetic tRNAs) or 10 µg of total RNA in 1× methylation buffer (50 mM Tris/HCl pH 7.0, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT) and 1.7 µM [3H]-SAM (Hartmann), 1 unit/mL RNasin (Promega) were incubated at 37°C for 2 h. Addition of proteinase K for 30 min stopped the reaction. The samples were separated on a 12% denaturing (7 M urea) polyacrylamide gel, stained with ethidium bromide, fixed and immersed for 1 h in amplify solution (Amersham). After drying, the gel was exposed to an X-ray film for between 16 h and 2 wk at −80°C.
Cell fractionation and Western blot
Cell fractionation was performed essentially as described previously (Haag et al. 2015). In brief, cells were harvested in 10 mM Tris pH 8.4, 140 mM NaCl, 1.5 mM MgCl2, 0.5 mM ETDA, 0.5 mM DTT, 0.5% (v/v) NP-40 and centrifuged at 100,000g for 10 min at 4°C. The supernatant (cytoplasmic extract) was collected and the pellet containing the nuclei was washed and directly resuspended in 5× SDS loading buffer (30 mM Tris–HCL pH 6.8, 10% SDS, 50% glycerol, 5% β-mercaptoethanol, bromophenolblue). Proteins were separated on 12% SDS gels and transferred to nitrocellulose membrane. Anti-FLAG (Sigma), anti-tubulin (Sigma), and anti-UTP14A (Proteintech) antibodies were used according to the suppliers’ instructions.
Microscopy
Flp-In T-Rex 293 cells stably expressing GFP-NSUN6 or NSUN6-GFP under a tetracycline-inducible promoter were seeded on coverslips. Expression was induced by 1 µg/mL tetracycline for 16–24 h. Cells were fixed using 4% paraformaldehyde in phosphate buffer saline (PBS) for 20 min at room temperature (RT) and permeabilized by 0.1% triton in PBS for 15 min. Cells were blocked by 10% FCS and 0.1% triton in PBS for 1 h at RT. Cells were incubated with primary antibodies against PCM1 (Cell Signaling), γ-tubulin (Bärenz et al. 2013), GM130 (MBL), and GOPC (Sigma-Aldrich) diluted in 10% FCS in PBS overnight at 4°C. Alexa Fluor 594 (Life Technologies) secondary antibodies were incubated with the cells for 2 h at RT. Coverslips were mounted on microscope slides using Vectashield mounting medium (Vector Labs). Fluorescence was examined using a confocal microscope and representative images are shown.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Blanche Schwappach and Oliver Gruss for sharing antibodies and Katherine Sloan for comments on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SPP 1784: BO3442/2-1 to M.T.B.; HO4436/2-1 to C.H.) and the Faculty of Medicine, Georg-August-University Göttingen (M.T.B. and “Startförderung” to S.H.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.051524.115.
Freely available online through the RNA Open Access option.
REFERENCES
- Banerjee S, Fisher O, Lohia A, Ankri S. 2005. Entamoeba histolytica DNA methyltransferase (Ehmeth) is a nuclear matrix protein that binds EhMRS2, a DNA that includes a scaffold/matrix attachment region (S/MAR). Mol Biochem Parasitol 139: 91–97. [DOI] [PubMed] [Google Scholar]
- Bärenz F, Inoue D, Yokoyama H, Tegha-Dunghu J, Freiss S, Draeger S, Mayilo D, Cado I, Merker S, Klinger M, et al. 2013. The centriolar satellite protein SSX2IP promotes centrosome maturation. J Cell Biol 202: 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M, Müeller S, Nellen W, Jurkowski TP, Jeltsch A, Ehrenhofer-Murray AE. 2012. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res 40: 11648–11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betat H, Long Y, Jackman JE, Mörl M. 2014. From end to end: tRNA editing at 5′- and 3′-terminal positions. Int J Mol Sci 15: 23975–23998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV, Iglesias A, Zhan Z, Fojo T. 1998. Like p53, the proliferation-associated protein p120 accumulates in human cancer cells following exposure to anticancer drugs. Biochem Biophys Res Commun 244: 368–373. [DOI] [PubMed] [Google Scholar]
- Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. 2014. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 33: 2020–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocker T, Bittinger A, Wieland W, Buettner R, Fauser G, Hofstaedter F, Rüschoff J. 1995. In vitro and ex vivo expression of nucleolar proteins B23 and p120 in benign and malignant epithelial lesions of the prostate. Mod Pathol 8: 226–231. [PubMed] [Google Scholar]
- Bohnsack MT, Tollervey D, Granneman S. 2012. Identification of RNA helicase target sites by UV cross-linking and analysis of cDNA. Methods Enzymol 511: 275–288. [DOI] [PubMed] [Google Scholar]
- Botticelli AR, Casali AM, Botticelli L, Zaffe D. 1998. Immunohistochemical detection of cell-cycle associated markers on paraffin embedded and formalin fixed needle biopsies of prostate cancer: correlation of p120 protein expression with AgNOR, PCNA/cyclin, Ki-67/MIB1 proliferation-scores and Gleason gradings. Eur J Histochem 42: 41–48. [PubMed] [Google Scholar]
- Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. 2006. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res 34: 6034–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cámara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, et al. 2011. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab 13: 527–539. [DOI] [PubMed] [Google Scholar]
- Cerrudo CS, Ghiringhelli PD, Gomez DE. 2014. Protein universe containing a PUA RNA-binding domain. FEBS J 281: 74–87. [DOI] [PubMed] [Google Scholar]
- Chen Y, Beck A, Davenport C, Chen Y, Shattuck D, Tavtigian SV. 2005. Characterization of TRZ1, a yeast homolog of the human candidate prostate cancer susceptibility gene ELAC2 encoding tRNase Z. BMC Mol Biol 6: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll A, Grzeschik KH. 2001. Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams-Beuren syndrome. Cytogenet Cell Genet 95: 20–27. [DOI] [PubMed] [Google Scholar]
- Fonagy A, Swiderski C, Ostrovsky AM, Bolton WE, Freeman JW. 1994. Effect of nucleolar P120 expression level on the proliferation capacity of breast cancer cells. Cancer Res 54: 1859–1864. [PubMed] [Google Scholar]
- Fonagy A, Swiderski C, Freeman JW. 1995. Altered transcription control is responsible for the increased level of proliferation-associated P120 in rapidly growing breast carcinoma. Int J Cancer 60: 407–412. [DOI] [PubMed] [Google Scholar]
- Freeman JW, McGrath P, Bondada V, Selliah N, Ownby H, Maloney T, Busch RK, Busch H. 1991. Prognostic significance of proliferation associated nucleolar antigen P120 in human breast carcinoma. Cancer Res 51: 1973–1978. [PubMed] [Google Scholar]
- Frye M, Watt FM. 2006. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol 16: 971–981. [DOI] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. 2006. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311: 395–398. [DOI] [PubMed] [Google Scholar]
- Haag S, Kretschmer J, Bohnsack MT. 2015. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA 21: 180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T, Marquez B, Suarez S, Schimenti J. 2007. Sperm motility defects and infertility in male mice with a mutation in Nsun7, a member of the Sun domain-containing family of putative RNA methyltransferases. Biol Reprod 77: 376–382. [DOI] [PubMed] [Google Scholar]
- Hopper AK. 2013. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 194: 43–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H. 2014. Methylated nucleosides in tRNA and tRNA methyltransferases. Front Genet 5: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Benavente SB, Nascimento E, Dragoni I, Kurowski A, Gillich A, Humphreys P, Frye M. 2009. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J Cell Biol 186: 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, et al. 2013a. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep 4: 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Tuorto F, Menon S, Blanco S, Cox C, Flores JV, Watt S, Kudo NR, Lyko F, Frye M. 2013b. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol Cell Biol 33: 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani R, Nureki O, Fukai S, Kijimoto T, Nameki N, Watanabe M, Kondo H, Sekine M, Okada N, Nishimura S, et al. 2002. Crystal structure of archaeosine tRNA-guanine transglycosylase. J Mol Biol 318: 665–677. [DOI] [PubMed] [Google Scholar]
- Job B, Bernheim A, Beau-Faller M, Camilleri-Broët S, Girard P, Hofman P, Mazières J, Toujani S, Lacroix L, Laffaire J, et al. 2010. Genomic aberrations in lung adenocarcinoma in never smokers. PLoS One 5: e15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, Jeltsch A. 2008. Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. RNA 14: 1663–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V, Cairns BR. 2013. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol 31: 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosronezhad N, Colagar AH, Jorsarayi SG. 2014. T26248G-transversion mutation in exon7 of the putative methyltransferase Nsun7 gene causes a change in protein folding associated with reduced sperm motility in asthenospermic men. Reprod Fertil Dev 10.1071/RD13371. [DOI] [PubMed] [Google Scholar]
- King MY, Redman KL. 2002. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry 41: 11218–11225. [DOI] [PubMed] [Google Scholar]
- Kuhlmann M, Borisova BE, Kaller M, Larsson P, Stach D, Na J, Eichinger L, Lyko F, Ambros V, Soderbom F, et al. 2005. Silencing of retrotransposons in Dictyostelium by DNA methylation and RNAi. Nucleic Acids Res 33: 6405–6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merla G, Ucla C, Guipponi M, Reymond A. 2002. Identification of additional transcripts in the Williams-Beuren syndrome critical region. Hum Genet 110: 429–438. [DOI] [PubMed] [Google Scholar]
- Metodiev MD, Spåhr H, Loguercio Polosa P, Meharg C, Becker C, Altmueller J, Habermann B, Larsson NG, Ruzzenente B. 2014. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet 10: e1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Helm M. 2011. RNA nucleotide methylation. Wiley Interdiscip Rev RNA 2: 611–631. [DOI] [PubMed] [Google Scholar]
- Mueller S, Windhof IM, Maximov V, Jurkowski T, Jeltsch A, Förstner KU, Sharma CM, Gräf R, Nellen W. 2013. Target recognition, RNA methylation activity and transcriptional regulation of the Dictyostelium discoideum Dnmt2-homologue (DnmA). Nucleic Acids Res 41: 8615–8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson L, Deutscher MP. 2000. Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J Biol Chem 275: 31559–31562. [DOI] [PubMed] [Google Scholar]
- Ohtsubo C, Shiokawa D, Kodama M, Gaiddon C, Nakagama H, Jochemsen AG, Taya Y, Okamoto K. 2009. Cytoplasmic tethering is involved in synergistic inhibition of p53 by Mdmx and Mdm2. Cancer Sci 100: 1291–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. 2004. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell 117: 311–321. [DOI] [PubMed] [Google Scholar]
- Phalke S, Nickel O, Walluscheck D, Hortig F, Onorati MC, Reuter G. 2009. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat Genet 41: 696–702. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddatz G, Guzzardo PM, Olova N, Fantappié MR, Rampp M, Schaefer M, Reik W, Hannon GJ, Lyko F. 2013. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci 110: 8627–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert AS, Thurlow DL, Mörl M. 2001. A eubacterial origin for the human tRNA nucleotidyltransferase? Biol Chem 382: 1431–1438. [DOI] [PubMed] [Google Scholar]
- Rieder R, Hobartner C, Micura R. 2009. Enzymatic ligation strategies for the preparation of purine riboswitches with site-specific chemical modifications. Methods Mol Biol 540: 15–24. [DOI] [PubMed] [Google Scholar]
- Rossmanith W. 2011. Localization of human RNase Z isoforms: dual nuclear/mitochondrial targeting of the ELAC2 gene product by alternative translation initiation. PLoS One 6: e19152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi DV, Norment A, Garrett CE. 1984. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci 81: 6993–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Lyko F. 2010. Lack of evidence for DNA methylation of Invader4 retroelements in Drosophila and implications for Dnmt2-mediated epigenetic regulation. Nat Genet 42: 920–921. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Steringer JP, Lyko F. 2008. The Drosophila cytosine-5 methyltransferase Dnmt2 is associated with the nuclear matrix and can access DNA during mitosis. PLoS ONE 3: e1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. 2010. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24: 1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33: W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, Calle-Perez A, Pircher A, Gerstl MP, Pfeifenberger S, et al. 2015. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun 6: 6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert C. 2009. The genomic basis of the Williams-Beuren syndrome. Cell Mol Life Sci 66: 1178–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam R, Aklujkar M, Schaefer M, Reinhardt R, Nickel O, Reuter G, Lovley DR, Ehrenhofer-Murray A, Nellen W, Ankri S, et al. 2014. The Dnmt2 RNA methyltransferase homolog of Geobacter sulfurreducens specifically methylates tRNA-Glu. Nucleic Acids Res 42: 6487–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Yang J, Watzinger P, Kotter P, Entian KD. 2013. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res 41: 9062–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronek E, Grzechnik P, Späth B, Marchfelder A, Kufel J. 2014. tRNA 3′ processing in yeast involves tRNase Z, Rex1, and Rrp6. RNA 20: 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Bohnsack MT, Watkins NJ. 2013. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep 5: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Leisegang MS, Doebele C, Ramirez AS, Simm S, Safferthal C, Kretschmer J, Schorge T, Markoutsa S, Haag S, et al. 2015. The association of late-acting snoRNPs with human pre-ribosomal complexes requires the RNA helicase DDX21. Nucleic Acids Res 43: 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. 2012. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 40: 5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. 2013. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell 51: 539–551. [DOI] [PubMed] [Google Scholar]
- Takaku H, Minagawa A, Takagi M, Nashimoto M. 2003. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res 31: 2272–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovy A, Hofmann B, Helm M, Ankri S. 2010. In vitro tRNA methylation assay with the Entamoeba histolytica DNA and tRNA methyltransferase Dnmt2 (Ehmeth) enzyme. J Vis Exp 44: 2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. 2012. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19: 900–905. [DOI] [PubMed] [Google Scholar]
- Wang G, Amanai K, Wang B, Jiang J. 2000. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev 14: 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Bohnsack MT. 2012. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 3: 397–414. [DOI] [PubMed] [Google Scholar]
- Weis BL, Missbach S, Marzi J, Bohnsack MT, Schleiff E. 2014. The 60S associated ribosome biogenesis factor LSG1-2 is required for 40S maturation in Arabidopsis thaliana. Plant J 80: 1043–1056. [DOI] [PubMed] [Google Scholar]
- Wu JC, Santi DV. 1987. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem 262: 4778–4786. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.