Abstract
Genes coding for members of the Sm-like (LSm) protein family are conserved through evolution from prokaryotes to humans. These proteins have been described as forming homo- or heterocomplexes implicated in a broad range of RNA-related functions. To date, the nuclear LSm2-8 and the cytoplasmic LSm1-7 heteroheptamers are the best characterized complexes in eukaryotes. Through a comprehensive functional study of the LSm family members, we found that lsm-1 and lsm-3 are not essential for C. elegans viability, but their perturbation, by RNAi or mutations, produces defects in development, reproduction, and motility. We further investigated the function of lsm-1, which encodes the distinctive protein of the cytoplasmic complex. RNA-seq analysis of lsm-1 mutants suggests that they have impaired Insulin/IGF-1 signaling (IIS), which is conserved in metazoans and involved in the response to various types of stress through the action of the FOXO transcription factor DAF-16. Further analysis using a DAF-16::GFP reporter indicated that heat stress-induced translocation of DAF-16 to the nuclei is dependent on lsm-1. Consistent with this, we observed that lsm-1 mutants display heightened sensitivity to thermal stress and starvation, while overexpression of lsm-1 has the opposite effect. We also observed that under stress, cytoplasmic LSm proteins aggregate into granules in an LSM-1-dependent manner. Moreover, we found that lsm-1 and lsm-3 are required for other processes regulated by the IIS pathway, such as aging and pathogen resistance.
Keywords: Caenorhabditis elegans; stress response; LSM, daf-16; P bodies; stress granules
INTRODUCTION
The presence of a domain forming a tertiary structure known as “Sm-fold” is the common signature of the large Sm/LSm(Sm-like) protein family. Genes encoding Sm/LSm proteins exist in Archaea, Bacteria, and Eukaryotes (Mura et al. 2013; Weichenrieder 2014). The Sm-fold mediates the interaction between Sm/LSm proteins in order to make multimeric complexes involved in many aspects of RNA metabolism (Wilusz and Wilusz 2013). While eukaryotic members of the Sm family form protein complexes that are components of different snRNPs in the spliceosome (U1, U2, U4, U5, U11, and U12), the LSm proteins have expanded specialized RNA-related functions including splicing, nuclear RNA processing, and messenger RNA decay (Tharun 2009; Veretnik et al. 2009).
Eukaryotic LSm proteins are distributed in two distinct LSm complexes, the nuclear LSm2-8 and the cytoplasmic LSm1-7 (Tharun 2009). Therefore, LSm2 to LSm7 are common subunits of the two complexes, while Lsm8 and Lsm1 are specific for nuclear and cytoplasmic compartments, respectively. The nuclear complex binds to U6 snRNA in the U6 snRNP and is involved in splicing, whereas the cytoplasmic complex has been described as an activator of the decapping step in the 5′–3′ mRNA decay pathway in P bodies (Tharun et al. 2000; Parker and Sheth 2007).
Besides the above-mentioned canonical LSm complexes, other heteromeric rings have been characterized in yeast and vertebrate cells pointing toward an expanded catalog of LSm functions in the modulation of RNA–RNA and RNA–protein interactions (Tomasevic and Peculis 2002; Pillai et al. 2003; Fernandez et al. 2004). Moreover, by doing RNAi against lsm genes in diverse genetic backgrounds, we and others have observed a greater functional complexity than expected from the two described heteroheptameric complexes (Ceron et al. 2007). The goal of the present study is to explore the functions of LSm proteins in Caenorhabditis elegans and therefore provide insights into their roles in the physiology and development of multicellular animals.
A phylogenetic analysis based on protein sequences revealed that the C. elegans genome contains all eight genes (lsm-1, lsm-2, lsm-3, lsm-4, lsm-5, lsm-6, lsm-7, and lsm-8) coding for small proteins (77–125 amino acids) of the canonical eukaryotic complexes, and three other genes (Y48G1C.9, K07A1.15, and C49H3.4) coding for small proteins with only Sm domains (Supplemental Fig. S1A). In addition to this set of lsm genes, other well-characterized and conserved Sm-like genes including edc-3 and car-1 encode larger proteins containing other functional domains (Supplemental Fig. S1A; Squirrell et al. 2006; Tritschler et al. 2007).
The study of LSm proteins in multicellular organisms has biomedical relevance since the overexpression of LSm1/CaSm (cancer-associated Sm-like) has been associated with malignant development in diverse types of human cancers (Streicher et al. 2007; Watson et al. 2008), although the causal oncogenic mechanism of LSm1 in tumor cells is unknown.
RESULTS AND DISCUSSION
LSM-1 and LSM-3 are not essential for C. elegans viability but contribute to the regulation of development, reproduction, and motility
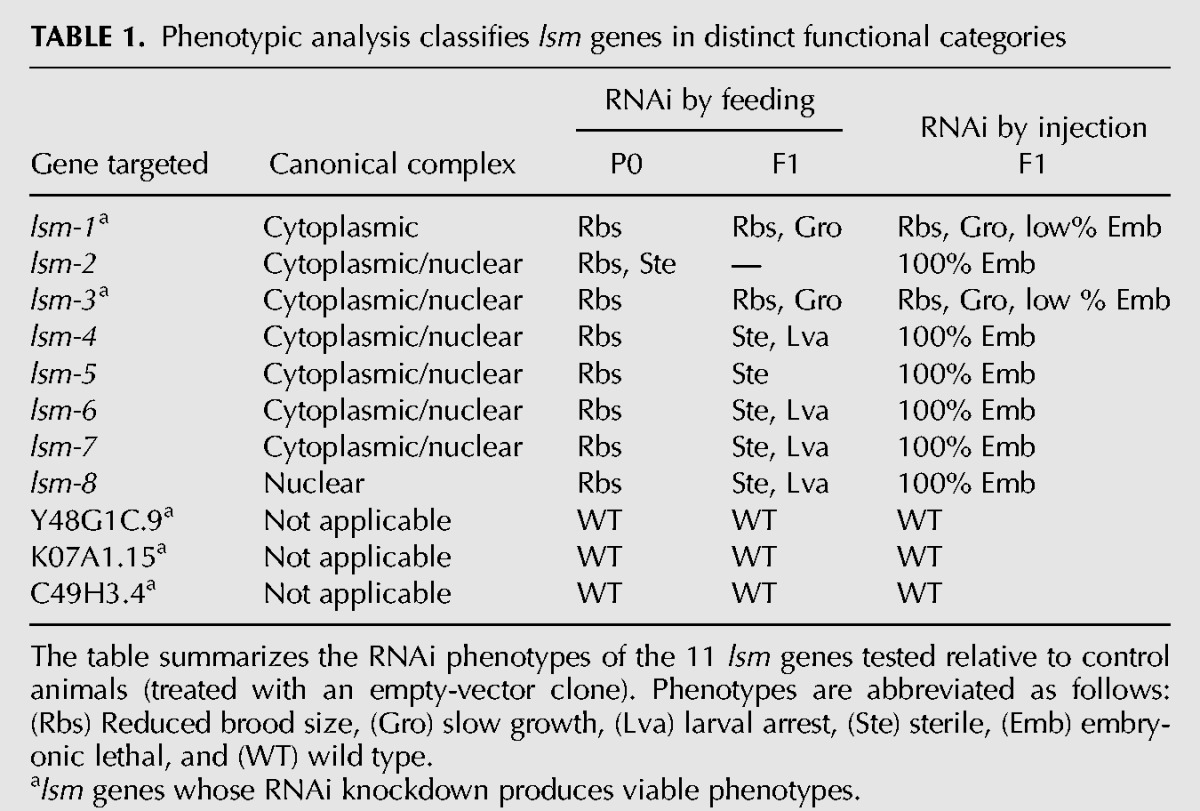
To shed light on the roles of the LSm complexes in C. elegans, we initiated functional studies of the components of the two canonical complexes as well as for Y48G1C.9, K07A1.15, and C49H3.4. First, we performed RNAi using both feeding and injection methods (Table 1). These experiments showed that (i) the three genes encoding LSm proteins that are not present in the canonical complexes are not required for the viability of the animals; and (ii) there is a heterogeneous phenotypic signature for the core lsm genes: lsm-2, lsm-4, lsm-5, lsm-6, lsm-7, and lsm-8 are essential, while lsm-1 and lsm-3 are not. These phenotypes are consistent with genetic data in yeast with the exceptions of lsm6 and lsm7, which are not essential in yeast (Supplemental Table S1); and lsm3, which is essential (Mayes et al. 1999; Salgado-Garrido et al. 1999). Therefore, lsm1/lsm-1 is the only nonessential member in both organisms.
TABLE 1.
Phenotypic analysis classifies lsm genes in distinct functional categories
We validated our RNAi results by analyzing mutants for lsm-1 and lsm-3. Both these mutants contain deletions that lack almost half of their transcripts, disrupting the Sm-like domain of the proteins (Fig. 1A). Both deletions are putatively functional null alleles since the corresponding dsRNAs delivered by microinjection did not further modify the mutant phenotype (Supplemental Fig. S1B). lsm-1(tm3585) and lsm-3(tm5166) mutant strains are viable but display pleiotropic phenotypes. Both mutations cause a low incidence of larval arrest, adult lethality, and embryonic lethality (Table 2). Moreover, lsm-1(tm3585) and lsm-3(tm5166) both exhibit reduced locomotor activity (Fig. 1B), are small compared with wild-type worms (Fig. 1C) and result in reduced brood sizes (Rbs) at 15°C. This Rbs phenotype is more pronounced at higher temperatures (Fig. 1D). We detected a reduction in the number of germ cells in adult lsm-1 worms compared with wild-type worms (Supplemental Fig. S1C), which is the possible cause of the reduced fertility. Low brood size was also observed for worms with mutations in other P body components such as homologs of the decapping enzyme DCAP-2/Dcp2 and the translation repression related protein CGH-1/Dhh1 (Supplemental Fig. S1D).
FIGURE 1.
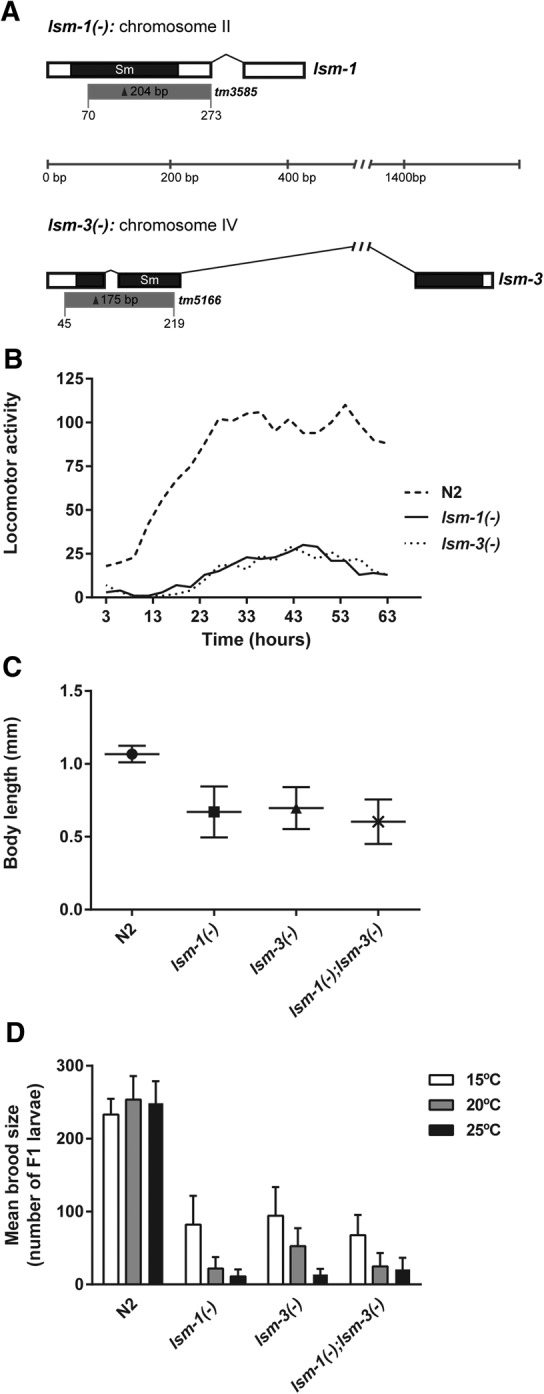
Characterization of lsm-1 and lsm-3 mutants. (A) Gene structures of lsm-1 and lsm-3. White boxes represent exons. Connecting lines represent introns. Black areas indicate the regions encoding the conserved Sm domain. Gray boxes represent regions deleted by lsm-1(tm3585) and lsm-3(tm5166) alleles; (bp) base pair scale. (B) Measure of animal motility using an automated locomotor tracking system (Simonetta and Golombek 2007). Locomotor activity plots showing the activity of L4 worms at 20°C (1 worm/well in a 96-well microplate, eight wells per genotype were analyzed). Each dot corresponds to the mean activity (measured in bins/hour) of 3 h intervals. (C) Graph representing the body length of young adults (grown for 48 h at 25°C from L1 stage). Mean and standard deviations are plotted (n ≥ 100 worms per genotype tested). (D) Brood sizes of wild type (N2) and the indicated mutant strains. Bars represent the standard deviation among individuals (n ≥ 20).
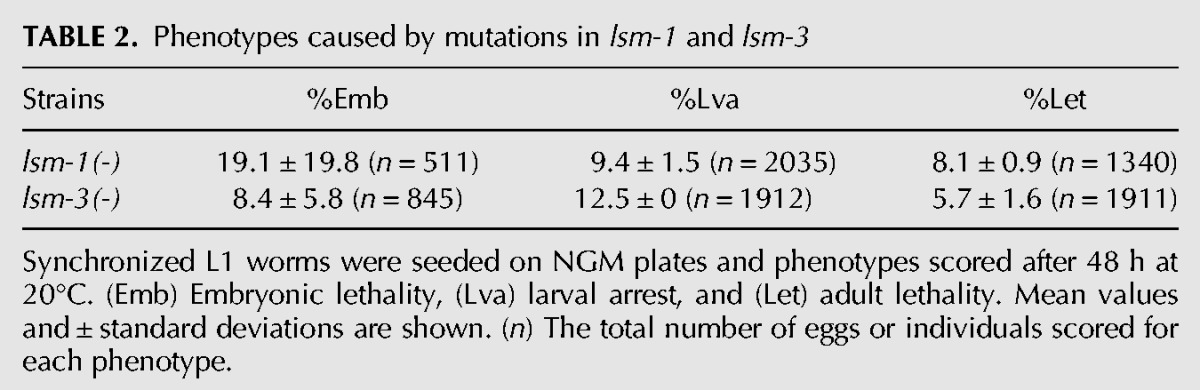
TABLE 2.
Phenotypes caused by mutations in lsm-1 and lsm-3
We generated a transgenic strain containing integrated copies of a LSM-1::GFP reporter with the lsm-1(tm3585). The reporter rescued the developmental and Rbs phenotypes (Supplemental Fig. S1E), indicating that the lsm-1 deletion is directly responsible for the observed phenotypes and demonstrated that our integrated LSM-1::GFP reporter is functional.
In yeast, lsm3 is essential for viability and necessary for correct splicing (Mayes et al. 1999). Since spliceosome core components are expected to be essential in C. elegans (Kamath et al. 2003; Rual et al. 2004; Kerins et al. 2010), the viability of lsm-3 mutants argues against a central role for LSM-3 in splicing. To test for any relevance of lsm-3 in this process, we generated a strain carrying a GFP reporter for constitutive splicing and the lsm-3 deletion. These animals displayed normal GFP expression in the intestinal nuclei, indicating that LSM-3 is not required for constitutive splicing (Supplemental Fig. S2A).
Given that neither lsm-1 nor lsm-3 are necessary for the survival of the animal, we tested to see if these genes have redundant functions by making a double homozygous mutant strain. The lack of a synthetic phenotype (Fig. 1C,D) and the fact that lsm-3 and lsm-1 mutants phenocopy each other indicate that these genes are not redundant but might participate in the same cellular processes during reproduction and development.
We also tested for additional synthetic interactions between the nonessential lsm genes in C. elegans by microinjecting combinations of diverse dsRNAs (Supplemental Fig. S1F). Since we did not observe any synthetic interaction, we concluded that C. elegans’ LSm family does not function as in Arabidopsis, where functional redundancies among Lsm proteins are evident (Perea-Resa et al. 2012).
In summary, our analysis showed that lsm-1 and lsm-3 are the only nonessential genes coding for members of the canonical LSm complexes, nevertheless, they are required for normal C. elegans development and health.
Expression of lsm genes in C. elegans
In addition to the apparent functional diversity of the lsm genes in C. elegans, we wanted to know if differences also exist at the regulatory level. The features of lsm genes in the C. elegans genome indicate heterogeneity in terms of regulatory regions because lsm genes present distinct UTR sequences and four out of the eleven genes are located in predicted operons (Supplemental Fig. S3A). We produced reporter constructs by PCR fusion (Hobert 2002) to generate promoter::GFP::unc-54_3′-UTR transgenic lines for the 11 lsm genes studied in this report (Supplemental Table S3). All promoters reported expression except for K07A1.15. We analyzed fluorescence intensities for several identifiers in C. elegans’ anatomy and observed overlapping but distinct gene expression patterns across family members (Supplemental Fig. S3B). However, these transcriptional reporters provide insufficient information to draw major conclusions because they do not contain all the regulatory sequences. As an example of the importance of other regulatory regions, a translational reporter for lsm-4 in a fosmid context (including UTRs and introns) was ubiquitously expressed although with apparent varying levels depending on the cell type (Supplemental Fig. S3C).
However, this localizome study showed that internal promoters can drive the expression of lsm genes located within different purported operons (as lsm-3 and lsm-8). Since both SL1 and SL2 splice leaders have been detected upstream of their messenger RNA (Allen et al. 2011), the activity of these internal promoters may not fully capture the endogenous expression pattern.
lsm-1 and lsm-3 are required for various stress responses
To gain insight into LSM-1 functions, the LSm protein with the highest biomedical relevance, we compared the transcriptomes of L3 (where the germline is not yet fully developed) lsm-1(tm3585) and wild-type animals (Supplemental Table S4). RNA-seq data suggest that genes related to the Insulin/IGF-1 signaling (IIS) pathway are mis-regulated in lsm-1(tm3585) mutants (Supplemental Table S4; McElwee et al. 2003; Murphy et al. 2003; Liu et al. 2004; Halaschek-Wiener et al. 2005; Oh et al. 2006; Pinkston-Gosse and Kenyon 2007; Lee et al. 2009; Schuster et al. 2010). The IIS pathway has been associated with immune response and stress protection, processes interconnected and regulated by DAF-2, which is the C. elegans ortholog of the insulin-like growth factor-1 (IGF-1) receptor (Troemel et al. 2006; Singh and Aballay 2009; Murphy and Hu 2013). In C. elegans, DAF-2 activity keeps the FOXO transcription factor DAF-16 in the cytoplasm. However, inactivation of DAF-2 signaling allows DAF-16 to translocate to the nucleus, thus inducing stress response at the transcriptional level (Yen et al. 2011).
Remarkably, among the top 24 up-regulated genes (fold change >3, Supplemental Table S4) in lsm-1(tm3585) mutants, we found four genes that are normally down-regulated by the IIS effector DAF-16 (C32H11.4, C32H11.9, dod-21, and dod-24) (Murphy et al. 2003). Moreover, pud-1.1 and pud-2.2, which are up-regulated in daf-2 mutants (Dong et al. 2007), are among the top 17 down-regulated genes (fold change < −3, Supplemental Table S4) in lsm-1(tm3585) mutants. Therefore, our transcriptome data point toward a DAF-16 deficiency in lsm-1(tm3585) mutants under normal conditions.
To further investigate the functional relationship between LSM-1 and the IIS pathway, we studied the stress-induced nuclear translocation dynamics of DAF-16 in the lsm-1(tm3585) mutant background. Interestingly, we observed that upon heat stress the translocation of DAF-16 to the nucleus is impaired in lsm-1(tm3585) mutants (Fig. 2A,B), a defect previously associated with diminished stress resistance (Lin et al. 2001). We observed a delay in the DAF-16 nuclear translocation although most of DAF-16 is eventually relocated to the nuclei. Such an effect is a hallmark of mutations that affect the correct function of the IIS pathway (Chiang et al. 2012). The DAF-16::GFP reporter that we used and lsm-3 are located in the same chromosome, making the generation of a genetic hybrid difficult. Thus to test the impact of lsm-3 on DAF-16 nuclear relocalization kinetics, we injected lsm-3 dsRNA and lsm-1 dsRNA as positive controls in a DAF-16::GFP strain. In both cases, we observed a defect in DAF-16 nuclear translocation upon heat shock (Supplemental Fig. S2B).
FIGURE 2.
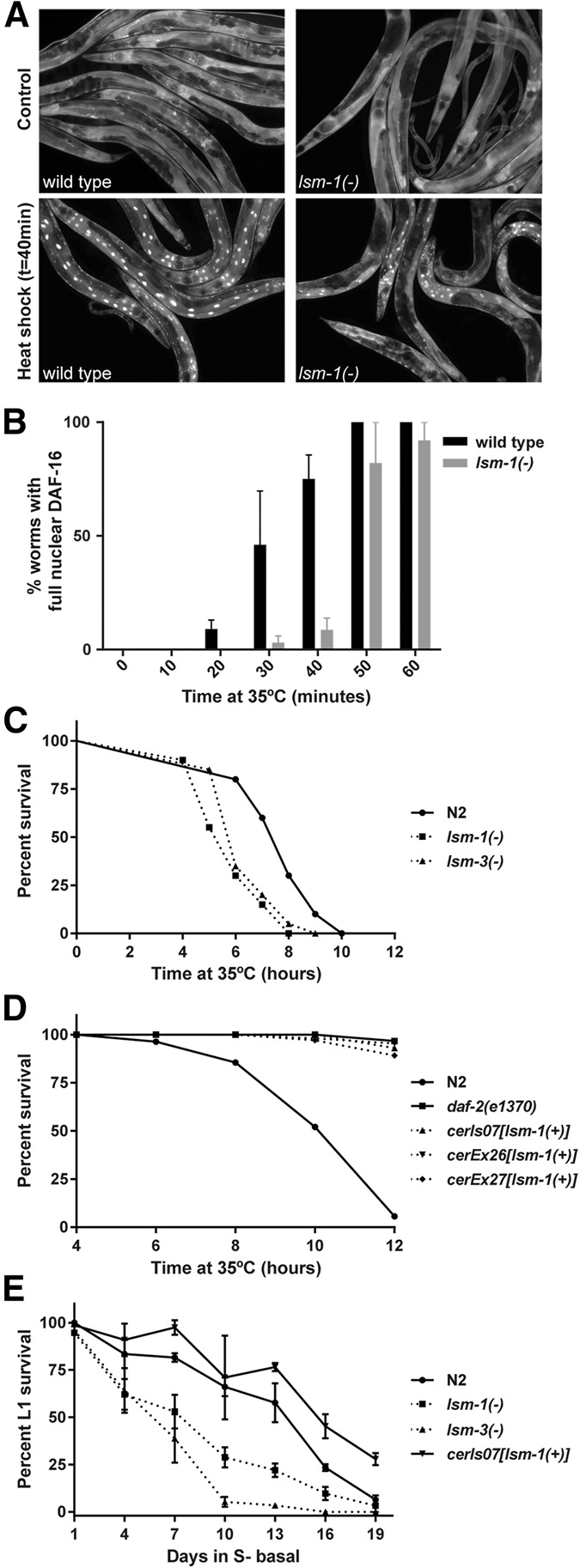
lsm-1 promotes DAF-16 nuclear localization upon stress and influences stress responses. (A) lsm-1 is required for proper stress-induced DAF-16 nuclear translocation. Fluorescence images of DAF-16::GFP in wild-type (N2) and lsm-1 mutant animals grown at 25°C, and after a 40 min heat shock at 35°C. (B) Time-course analysis of DAF-16::GFP nuclear accumulation in response to heat stress. The histogram represents the average of percentages of worms with full nuclear DAF-16::GFP in three independent experiments (n ≥ 50 for each strain and time point). Error bars, SEM. (C) Representative graph of experiments (see Supplemental Fig. S4A) showing the significant reduced resistance to heat stress caused by mutations in lsm-1 and lsm-3 (P-value < 0.001). Young adult worms were incubated at 35°C and survival was scored every hour. (D) Representative graph of experiments (see Supplemental Fig. S4A) showing the significant resistance to heat stress caused by three distinct strains overexpressing lsm-1 (P-value < 0.001). daf-2(e1370) were used as positive control for heat stress resistance. Young adult worms were incubated at 35°C and survival was scored every 2 h. (E) lsm-1 and lsm-3 mutations reduce the survival of L1 larvae during starvation at 20°C. Ectopic expression of lsm-1 through the integrated reporter LSM-1::GFP displayed a significant survival to starvation compared with wild-type (N2) animals after 19 d (P-value < 0.001). Graph represents the mean percentage of survival and standard deviations from three experimental replicates (n ≥ 100 for each replicate and time point).
Consistent with our previous observations, we found that lsm-1(tm3585) and lsm-3(tm5166) mutants were sensitive to heat stress (Fig. 2C), while strains overexpressing lsm-1 displayed a significant thermoresistance, similar to that reported for daf-2(e1370) mutants where DAF-16 is constitutively nuclear and transcriptionally active (Fig. 2D; McColl et al. 2010). Furthermore, we also observed that lsm-1(tm3585) and lsm-3(tm5166) mutant L1 larvae were more sensitive to starvation than wild-type animals, whereas larvae with extra copies of lsm-1 had a higher survival rate than wild-type worms (Fig. 2E). All these results support the idea that lsm-1 and lsm-3 contribute to stress responses through the IIS pathway in C. elegans and that lsm-1 levels influence the robustness of these responses.
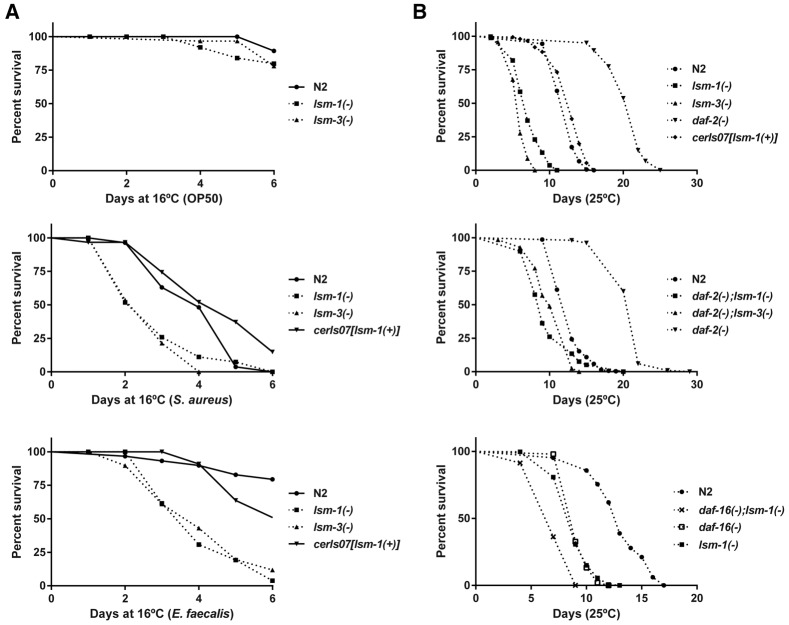
The IIS pathway also regulates resistance to bacterial infection and longevity (Evans et al. 2008). We observed that mutations in lsm-1 and lsm-3 cause heightened sensitivity to infection by certain pathogens (Fig. 3A) and have negative effects on lifespan (Fig. 3B). However, ectopic lsm-1 expression does not protect C. elegans from pathogens or extend its lifespan (Fig. 3). Moreover, the daf-2(m577) extended lifespan requires lsm-1 and lsm-3, and the short lifespan of daf-16(mu86) mutants is shortened by lsm-1(tm3585), suggesting that the harmful effect of the lsm-1 mutation on longevity is independent of the regulation of DAF-16 nuclear location.
FIGURE 3.
Mutations in lsm-1 and lsm-3 cause hypersensitivity to pathogen infection and affect IIS-induced longevity. (A) Representative graph of survival experiments (see Supplemental Fig. S4B) of wild-type (N2) and mutant strains on Escherichia coli OP50, Staphylococcus aureus, and Enterococcus faecalis (n = 30). No significant differences were observed after 6 d on OP50 between lsm-1 and lsm-3 mutants compared with wild-type (N2) animals. Differences between lsm-1 and lsm-3 mutants were significant compared with wild-type (N2) after pathogen infection (0.01 < P-value < 0.001 in all cases). (B) Representative graphs showing lifespan analysis (see Supplemental Fig. S4C) of wild-type (N2) and different mutant and transgenic strains. daf-2(m577) and daf-16(mu86) mutants were used as control for extended and shortened lifespans, respectively.
Redistribution of cytoplasmic LSm proteins in stress conditions
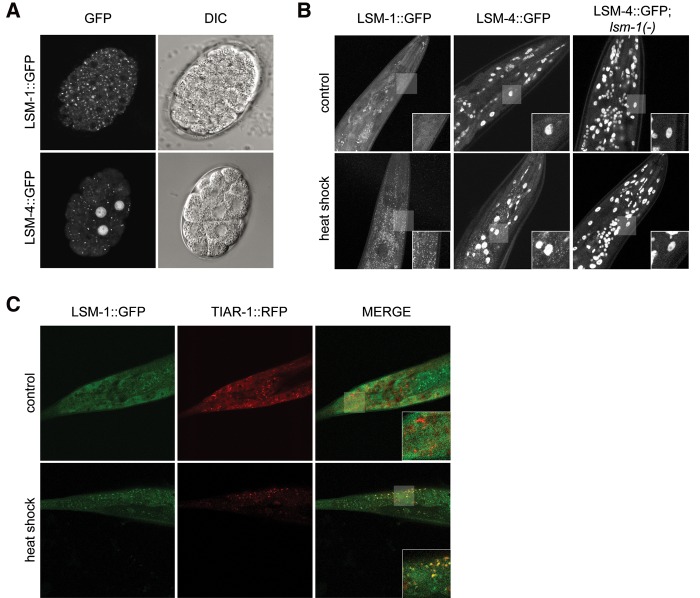
We generated integrated transgenic reporter lines for LSM-1 and LSM-4, which allowed the tracking of nuclear and cytoplasmic LSm complexes in the germline and early embryo. As previously observed in other systems (Ingelfinger et al. 2002; Spiller et al. 2007), LSM-1 was located exclusively in the cytoplasm whereas LSM-4 was expressed both in the nucleus and cytoplasm (Fig. 4A).
FIGURE 4.
LSM proteins accumulate in cytoplasmic foci in specific stages and conditions. (A) LSM proteins accumulate in cytoplasmic granules during embryogenesis. Confocal images of embryos expressing LSM-1::GFP in the cytoplasm and LSM-4::GFP in the nucleus and the cytoplasm. Right panels show the same embryos visualized under differential interference contrast (DIC). (B) LSm accumulation in cytoplasmic granules in adult cells is induced by heat stress (1 h at 35°C) (additional images in Supplemental Fig. S5) and is dependent on lsm-1. Confocal images (Z hyperstack) of the head of young adult worms expressing LSM-1::GFP and LSM-4::GFP in control (20°C) and after heat-shock. (C) LSM-1 granules colocalize with TIAR-1 granules under heat-stress conditions (1 h at 35°C). Confocal images of the tail of L4 worms coexpressing LSM-1::GFP and RFP::TIAR-1.
As previously reported in C. elegans, we observed constitutive cytoplasmic LSm granules in the early embryo in somatic blastomeres (Fig. 4A; Gallo et al. 2008). However, we found that in young adult animals the distribution of cytoplasmic LSm proteins was diffuse rather than located in aggregates (Fig. 4B). However, under heat stress, adult animals expressing LSM-1::GFP or LSM-4::GFP showed cytoplasmic GFP granules (Fig. 4B). Microscopic analysis of the LSM-4::GFP reporter in lsm-1 mutants indicated that the formation of cytoplasmic LSm granules after stress was dependent on the presence of LSM-1 (Fig. 4B).
In summary, while the formation of cytoplasmic LSM granules is constitutive in some embryonic cells, the accumulation of visible LSm granules in larvae and adults may occur upon stress only, in an LSM-1-dependent manner.
The described competition of LSm8 and LSm1 for the common components of the LSm complexes (LSm2-7) implies that nuclear and cytoplasmic functions are somehow interconnected, and such co-regulation should be carefully studied in multicellular organisms (Spiller et al. 2007; Novotny et al. 2012).
Cytoplasmic LSm granules: P-bodies or stress granules?
In yeast and humans, the LSm1-7 complex has been associated with P bodies (PBs) (Kedersha and Anderson 2007). PBs are dynamic cytoplasmic aggregates of proteins and RNA molecules that participate in diverse processes related to RNA metabolism such as translational repression and mRNA degradation (Sheth and Parker 2006; Parker and Sheth 2007; Buchan et al. 2010). In particular, the LSm1-7 complex has been implicated in the 5′–3′ mRNA decay machinery as a decapping activator of oligoadenilated mRNAs in P-bodies (Tharun 2009).
Stress granules (SGs) are another type of cytoplasmic aggregate, composed of nontranslating mRNAs and diverse proteins related to mRNA processing that form when translational initiation is impaired (for example, when cells are exposed to environmental stresses) (Buchan and Parker 2009).
PBs and SGs are dynamic structures that share some protein components and can physically interact (Kedersha and Anderson 2007; Buchan et al. 2010); however, the physiological roles of these aggregates are not well understood and the classification of proteins within one or another is mostly based on protein co-localization experiments with previously characterized proteins.
In the C. elegans embryo, it has been shown that LSM-1::GFP co-localizes with other PB components, but the presence of visible LSM-1::GFP granules is not required for mRNA degradation (Gallo et al. 2008). Comparing the localization of LSM-1 to the known P bodies component DCAP-2, which is the catalytic enzyme of the 5′–3′ decapping step in PBs, shows they are different. In developing C. elegans embryos, LSM-1 is present only in somatic blastomeres, whereas DCAP-2 localizes in P granules in germline blastomeres (Lall et al. 2005). Another PB component, the decapping enzyme DCAP-1, localizes in granules that are only weakly reduced upon lsm-1 RNAi inactivation (Sun et al. 2011). Moreover, in adult animals, although DCAP-1-positive granules have been described to increase in size with age (Sun et al. 2011), we did not observe this effect in LSM-1::GFP worms (not shown).
Distinct cytoplasmic aggregates can share many RNA-binding proteins and depending on cellular conditions, these factors can relocalize from one type of granule to another (Buchan 2014). Since LSM-1::GFP aggregates in stress conditions, LSM-1 may shuttle between PBs and SGs depending on physiological or environmental conditions. To confirm the aggregation of LSM-1 in SGs, we used an RFP-tagged TIAR-1 (TIA-1-related), which is a RNA-binding protein described as a SG component (Kedersha et al. 1999). We observed the co-localization of LSM-1::GFP and TIAR-1::RFP under heat stress (Fig. 4C), suggesting that LSM-1 can also accumulate in SG under specific conditions. In mammalian cells, LSm1 is mostly P body-specific in the absence of stress, although it has been observed associated with some types of stress granules (Kedersha and Anderson 2007).
Since the single bacterial LSm protein homolog Hfq is also required for stress responses in bacteria (Wilusz and Wilusz 2013), the capability of LSm proteins to chaperone RNA–protein and RNA–RNA interactions seems to be conserved through evolution as an adaptive response, distributing RNA molecules and regulating protein synthesis, to cope with adverse environmental conditions. Interestingly, such a response in C. elegans seems to be linked to the IIS pathway, also evolutionary conserved in Metazoa.
Concluding remarks
Through a comprehensive phenotypic analysis of the lsm family in C. elegans, we have uncovered the role of cytoplasmic LSm proteins in C. elegans stress response. We found that LSM-1 is required to establish a primary response to stress through two known mechanisms: (i) the translocation of DAF-16 to the nucleus, and (ii) the formation of cytoplasmic granules. To the best of our knowledge, this is the first time that a functional link has been observed between these two mechanisms in the stress response.
Regarding the link between LSm1 and cancer, it is tempting to establish a parallel between cancer cells and the conditions required in C. elegans for LSM-1 cytoplasmic aggregation, namely, rapid cell division in the embryo and environmental stress. Thus, we present C. elegans as an excellent model to further investigate the impact of cytoplasmic Lsm proteins in cellular metabolism and cancer.
MATERIALS AND METHODS
Strains
Caenorhabditis elegans strains were cultured and maintained using standard procedures (Stiernagle 2006; Porta-de-la-Riva et al. 2012). Bristol N2 was the wild-type strain, and the following alleles and transgenic strains were used: CER60: lsm-1(tm3585)II, CER59: lsm-3(tm5166)IV, CER36: lsm-1(tm3585)II, lsm-3(tm5166)IV, CB1370: daf-2(e1370)III, CF1038: daf-16(mu86)I, BL3466: inIs173[PNvitgfp], TJ356: zIs356[Pdaf-16::daf-16::gfp,rol-6(su1006)]IV, DR1567: daf-2(m577)III, CER154: lsm-1(tm3585)II;daf-2(m577)III, CER158: lsm-1(tm3585)II;daf-16(mu86)I, CER159: lsm-3(tm5166)IV;daf-2(m577)III, DG1701: cgh-1(tn691)III and RB1641: dcap-2(ok2023)IV.
We also generated the following transgenic strains: CER26: unc-119(ed3)III; cerEx26[Plsm-1::lsm-1::gfp::lsm-1_3′UTR, unc-119(+)], CER27: unc-119(ed3)III; cerEx27[Plsm-1::lsm-1::gfp::lsm-1_3′UTR, unc-119(+)], CER41: unc-119(ed3)III; cerIs02[Plsm-4::lsm-4::gfp::lsm-4_3′UTR, unc-119(+)], CER157: unc-119(ed3)III; cerIs07[Plsm-1::lsm-1::gfp::lsm-1_3′UTR, unc-119(+)], CER152: cerIs02; lsm-1(tm3585)II, CER155: zIs356IV;lsm-1(tm3585)II, CER129: inIs173[PNvitgfp];lsm-3(tm5166)IV, CER162: cerIs07;lsm-1(tm3585)II, CER166: cerIs07;cerEx42[Ptiar-1::RFP::tiar-1ORF::tiar-1 3′UTR+rol-6(+)].
Generation of transgenic animals
For the generation of lsm-1 and lsm-4 translational reporters, fosmid vectors containing a GFP-tagged version of these genes were requested from the Transgeneome resource (Sarov et al. 2012) and transformation was performed by bombardment with gold particles (Biolistic Helium Gun, Caenotec). unc-119(ed3) young adults were shot with 16 µg of the purified DNA of interest (Praitis et al. 2001).
For the co-localization experiments, CER157 animals expressing integrated copies of LSM-1::GFP were injected with a mix containing 80 ng/μL of a RFP::TIAR-1 construct (Rousakis et al. 2014) and 20 ng/μL of the linearized roller marker pRF4 [rol-6(su1006)].
Transcriptional reporters, described in the Supplemental Material, were constructed by PCR fusion (Hobert 2002).
RNAi
The RNAi clones used in this study were obtained from the ORFeome library (Rual et al. 2004) (lsm-1, lsm-3, lsm-5, lsm-7, C49H3.4) or the Ahringer library (Kamath et al. 2003) (lsm-2, lsm-4, lsm-6). We completed the lsm family RNAi clone collection by amplifying the cDNAs of lsm-8, Y48G1C.9, and K07A1.15, and cloning them into a L4440 vector by ligation after digestion with restriction enzymes. All RNAi clones were verified by sequencing. RNA-mediated interference (RNAi) by feeding was performed following standard conditions (Fontrodona et al. 2013), using a concentration of 3 mM IPTG on the RNAi plates. To induce RNAi by microinjection, specific dsRNAs were synthesized by using the MEGAscript T7 kit (Ambion). Young adult animals were injected with 1 μg/μL of the dsRNA of interest.
RNA-seq analyses
N2 wild-type and lsm-1(tm3585) L1 synchronized worms were grown and harvested at L3 stage (26 h at 25°C). Animals were washed with M9 buffer to remove bacteria, and pellet frozen in TRIzol. Total RNA purification was performed using the mirVana miRNA isolation kit (Ambion) followed by Ribosomal RNA depletion with the RiboMinus Eukaryote Kit (Invitrogen). RNA quality was verified in the Experion Bioanalyzer (Bio-Rad). We used an Illumina kit to make libraries that were run through a Genome Analyzer IIX Ultrasequencer (Illumina). Each sample produced ∼10 million reads that were processed using TopHat (Trapnell et al. 2009) to be mapped against the C. elegans genome (WS225). BAM files were analyzed with the SeqSolve NGS software (Integromics, S.L.) using a false discovery rate of 0.05, and filtering reads displaying multiple mapping sites. SeqSolve uses Cufflinks (Trapnell et al. 2010) and Cuffdiff (Trapnell et al. 2013) programs to perform differential gene expression analyses between samples (P-value < 0.005). Expression values were normalized in FPKM (fragments per kilobase of exon per million fragments mapped).
Stress assays
For thermotolerance assays, L4 animals grown at 16°C were transferred to plates seeded with OP50 bacteria and grown to day 1 of adulthood. Next, worms (n = 40) were transferred to two 6 cm plates without any food and incubated at 35°C. Viability was scored at several time points; death was determined by the lack of movement and/or pharyngeal pumping after prodding.
To quantify DAF-16::GFP subcellular localization, L4 worms grown at 16°C were shifted to 25°C until they reached day 1 adult stage. GFP was analyzed using an Axio Imager Z1 Zeiss microscope at 40× magnification before and after heat shock at 35°C. For heat shock time-course analyses, worms were scored for the presence or absence of GFP accumulation in the nuclei of somatic cells along the body every 10 min at 35°C. Animals were scored as having full nuclear GFP if DAF-16::GFP was observed in the nucleus of somatic cells homogeneously from head to tail.
L1 starvation assay was performed as previously described by Zhang et al. (2011). Briefly, adult worms were bleached and the resulting eggs were resuspended in 4–6 mL S-basal without cholesterol in 15-mL tubes. Egg prep was filtered using a 40 μm nylon cell strainer (BD Falcon) in order to remove debris and larger clumps of unhatched eggs derived from the bleaching. Tubes were incubated rotating at 20°C. To determine larval viability, 20-μL aliquots (∼100 worms) were placed every 3 d onto three 6-cm nematode growth medium (NGM) plates and survival rates were calculated.
For bacterial pathogen assays, L4 worms grown at 16°C (n = 10 for each genotype) were cultured in the presence of pathogenic bacteria as the sole food source, and the number of survivors was counted every day. Enterococcus faecalis OG1RF was grown in BHI with 40 μg/mL of gentamycin. Staphylococcus aureus NTCT8325 was grown in TSB with 10 μg/mL of nalidixic acid.
For all the stress assays, graphical representation of survival curves and Kaplan-Meier statistical analyses were made using GraphPad Prism 4.0 software (GraphPad Software Inc.) P-values were obtained applying the Mantel-Cox logrank test.
Lifespan experiments
A synchronized population of L1 larvae was grown at 15°C for 72 h until they reached L4 stage. At that point, worms were washed from the plate using sterile M9 buffer and transferred (at least 100 animals per experiment) onto NGM plates seeded with OP50 and 0.1 mg/mL 5-fluoro-2′-deoxyuridine (FUDR) to chemically inhibit reproduction and to eliminate any effects reproduction might have on longevity. The viability of the worms, cultured at 25°C was scored every two or three days. Animals that failed to respond to stimulation by touch were considered dead. Day 0 of adulthood was defined as the day that mid-to-late L4s were transferred to NGM-FUDR plates and maintained at 25°C. Survival curves were made with GraphPad Prism 4.0 software.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank the CGC (Caenorhabditis Genetics Center) and the Japanese C. elegans knockout consortium (National BioResource Project, NBRP) for providing strains; Dr. Popi Syntichaki for providing us with constructs; and Dr. Anne Royou and members from the Royou laboratory for confocal microscopy technical advice. This work was supported by a grant from the Instituto de Salud Carlos III (ISCIII) (Exp. PI12/01554). R.J. and D.B. were supported by a Natural Sciences and Engineering Research Council (NSERC) Canada grant. D.B. holds a Canada Research Chair. J.C. is a Miguel Servet Researcher (ISCIII). E.C. was supported with a CTP-AIRE fellowship from AGAUR (Generalitat de Catalunya) and a TRANSBIO SUDOE collaborative project.
Author contributions: E.C., D.D., and J.C. conceived and designed the experiments. E.C., D.A., M.P., A.B., I.E., M.S., L.F, F.J., K.R., and J.C. performed the experiments. E.C., D.D., and J.C. analyzed the data. R.J., D.B., M.M., and M.D. contributed reagents/materials/analysis tools. E.C., D.D., and J.C. wrote the paper.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.052324.115.
REFERENCES
- Allen MA, Hillier LW, Waterston RH, Blumenthal T. 2011. A global analysis of C. elegans trans-splicing. Genome Res 21: 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR. 2014. mRNP granules: assembly, function, and connections with disease. RNA Biol 11: 1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36: 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Nissan T, Parker R. 2010. Analyzing P-bodies and stress granules in Saccharomyces cerevisiae. Methods Enzymol 470: 619–640. [DOI] [PubMed] [Google Scholar]
- Ceron J, Rual J-F, Chandra A, Dupuy D, Vidal M, Van Den Heuvel S. 2007. Large-scale RNAi screens identify novel genes that interact with the C. elegans retinoblastoma pathway as well as splicing-related components with synMuv B activity. BMC Dev Biol 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang W-C, Tishkoff DX, Yang B, Wilson-Grady J, Yu X, Mazer T, Eckersdorff M, Gygi SP, Lombard DB, Hsu A-L. 2012. C. elegans SIRT6/7 homolog SIR-2.4 promotes DAF-16 relocalization and function during stress. PLoS Genet 8: e1002948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M-Q, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR. 2007. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317: 660–663. [DOI] [PubMed] [Google Scholar]
- Evans E, Chen WC, Tan M-W. 2008. The DAF-2 insulin-like signaling pathway independently regulates aging and immunity in C. elegans. Aging Cell 7: 879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CF, Pannone BK, Chen X, Fuchs G, Wolin SL. 2004. An Lsm2-Lsm7 complex in Saccharomyces cerevisiae associates with the small nucleolar RNA snR5. Mol Biol Cell 15: 2842–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontrodona L, Porta-de-la-Riva M, Morán T, Niu W, Díaz M, Aristizábal-Corrales D, Villanueva A, Schwartz S Jr, Reinke V, Cerón J. 2013. RSR-2, the Caenorhabditis elegans ortholog of human spliceosomal component SRm300/SRRM2, regulates development by influencing the transcriptional machinery. PLoS Genet 9: e1003543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo CM, Munro E, Rasoloson D, Merritt C, Seydoux G. 2008. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev Biol 323: 76–87. [DOI] [PubMed] [Google Scholar]
- Halaschek-Wiener J, Khattra JS, McKay S, Pouzyrev A, Stott JM, Yang GS, Holt RA, Jones SJM, Marra MA, Brooks-Wilson AR, et al. 2005. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res 15: 603–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. 2002. BioTechniques—PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32: 728–730. [DOI] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Lührmann R, Achsel T. 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8: 1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. 2007. Mammalian stress granules and processing bodies. Methods Enzymol 431: 61–81. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 α to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerins JA, Hanazawa M, Dorsett M, Schedl T. 2010. PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation versus meiotic development decision and germline sex determination in Caenorhabditis elegans. Dev Dyn 239: 1555–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S, Piano F, Davis RE. 2005. Caenorhabditis elegans decapping proteins: localization and functional analysis of Dcp1, Dcp2, and DcpS during embryogenesis. Mol Biol Cell 16: 5880–5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-J, Murphy CT, Kenyon C. 2009. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab 10: 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. 2001. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 28: 139–145. [DOI] [PubMed] [Google Scholar]
- Liu T, Zimmerman KK, Patterson GI. 2004. Regulation of signaling genes by TGFβ during entry into dauer diapause in C. elegans. BMC Dev Biol 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayes AE, Verdone L, Legrain P, Beggs JD. 1999. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J 18: 4321–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl G, Rogers AN, Alavez S, Hubbard AE, Melov S, Link CD, Bush AI, Kapahi P, Lithgow GJ. 2010. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab 12: 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J, Bubb K, Thomas JH. 2003. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2: 111–121. [DOI] [PubMed] [Google Scholar]
- Mura C, Randolph PS, Patterson J, Cozen AE. 2013. A structural and evolutionary perspective on Sm function Archaeal and eukaryotic homologs of Hfq. RNA Biol 10: 636–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, Hu PJ. 2013. Insulin/insulin-like growth factor signaling in C. elegans. In WormBook (ed. The C. elegans Research Community). http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283. [DOI] [PubMed] [Google Scholar]
- Novotny I, Podolská K, Blazíková M, Valásek LS, Svoboda P, Stanek D. 2012. Nuclear LSm8 affects number of cytoplasmic processing bodies via controlling cellular distribution of Like-Sm proteins. Mol Biol Cell 23: 3776–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. 2006. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet 38: 251–257. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. 2007. P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646. [DOI] [PubMed] [Google Scholar]
- Perea-Resa C, Hernández-Verdeja T, López-Cobollo R, del Mar Castellano M, Salinas J. 2012. LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell 24: 4930–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Grimmler M, Meister G, Will CL, Lührmann R, Fischer U, Schümperli D. 2003. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev 17: 2321–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkston-Gosse J, Kenyon C. 2007. DAF-16/FOXO targets genes that regulate tumor growth in Caenorhabditis elegans. Nat Genet 39: 1403–1409. [DOI] [PubMed] [Google Scholar]
- Porta-de-la-Riva M, Fontrodona L, Villanueva A, Cerón J. 2012. Basic Caenorhabditis elegans methods: synchronization and observation. J Vis Exp e4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousakis A, Vlanti A, Borbolis F, Roumelioti F, Kapetanou M, Syntichaki P. 2014. Diverse functions of mRNA metabolism factors in stress defense and aging of Caenorhabditis elegans. PLoS One 9: e103365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J-F, Ceron J, Koreth J, Hao T, Nicot A-S, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, et al. 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14: 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Séraphin B. 1999. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J 18: 3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M, Murray JI, Schanze K, Pozniakovski A, Niu W, Angermann K, Hasse S, Rupprecht M, Vinis E, Tinney M, et al. 2012. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150: 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster E, McElwee JJ, Tullet JMA, Doonan R, Matthijssens F, Reece-Hoyes JS, Hope IA, Vanfleteren JR, Thornton JM, Gems D. 2010. DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol 6: 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125: 1095–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta SH, Golombek DA. 2007. An automated tracking system for Caenorhabditis elegans locomotor behavior and circadian studies application. J Neurosci Methods 161: 273–280. [DOI] [PubMed] [Google Scholar]
- Singh V, Aballay A. 2009. Regulation of DAF-16-mediated innate immunity in Caenorhabditis elegans. J Biol Chem 284: 35580–35587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller MP, Reijns MAM, Beggs JD. 2007. Requirements for nuclear localization of the Lsm2-8p complex and competition between nuclear and cytoplasmic Lsm complexes. J Cell Sci 120: 4310–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squirrell JM, Eggers ZT, Luedke N, Saari B, Grimson A, Lyons GE, Anderson P, White JG. 2006. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol Biol Cell 17: 336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. 2006. Maintenance of C. elegans. In WormBook (ed. The C. elegans Research Community). http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streicher KL, Yang ZQ, Draghici S, Ethier SP. 2007. Transforming function of the LSM1 oncogene in human breast cancers with the 8p11-12 amplicon. Oncogene 26: 2104–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Yang P, Zhang Y, Bao X, Li J, Hou W, Yao X, Han J, Zhang H. 2011. A genome-wide RNAi screen identifies genes regulating the formation of P bodies in C. elegans and their functions in NMD and RNAi. Protein Cell 2: 918–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S. 2009. Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int Rev Cell Mol Biol 272: 149–189. [DOI] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404: 515–518. [DOI] [PubMed] [Google Scholar]
- Tomasevic N, Peculis BA. 2002. Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol Cell Biol 22: 4101–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Eulalio A, Truffault V, Hartmann MD, Helms S, Schmidt S, Coles M, Izaurralde E, Weichenrieder O. 2007. A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol Cell Biol 27: 8600–8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. 2006. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans ed. S. Kim. PLoS Genet 2: e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veretnik S, Wills C, Youkharibache P, Valas RE, Bourne PE. 2009. Sm/Lsm genes provide a glimpse into the early evolution of the spliceosome. PLoS Comput Biol 5: e1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PM, Miller SW, Fraig M, Cole DJ, Watson DK, Boylan AM. 2008. CaSm (LSm-1) overexpression in lung cancer and mesothelioma is required for transformed phenotypes. Am J Respir Cell Mol Biol 38: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenrieder O. 2014. RNA binding by Hfq and ring-forming (L)Sm proteins: a trade-off between optimal sequence readout and RNA backbone conformation. RNA Biol 11: 537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J. 2013. Lsm proteins and Hfq: life at the 3′ end. RNA Biol 10: 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Narasimhan SD, Tissenbaum HA. 2011. DAF-16/Forkhead box O transcription factor: many paths to a single Fork(head) in the road. Antioxid Redox Signal 14: 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zabinsky R, Teng Y, Cui M, Han M. 2011. microRNAs play critical roles in the survival and recovery of Caenorhabditis elegans from starvation-induced L1 diapause. Proc Natl Acad Sci 108: 17997–18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.