We report molecular characteristics of M. pneumoniae in respiratory specimens from children and adults hospitalized with CAP. The P1 type 1 genotype and MLVA type 4/5/7/2 predominated, but proportions of types differed between children and adults. Macrolide resistance was rare.
Keywords: community-acquired pneumonia, macrolide resistance, molecular epidemiology, Mycoplasma pneumonia
Abstract
Background. Mycoplasma pneumoniae is a common cause of community-acquired pneumonia (CAP). The molecular characteristics of M pneumoniae detected in patients hospitalized with CAP in the United States are poorly described.
Methods. We performed molecular characterization of M pneumoniae in nasopharyngeal/oropharyngeal swabs from children and adults hospitalized with CAP in the Centers for Disease Control and Prevention Etiology of Pneumonia in the Community (EPIC) study, including P1 typing, multilocus variable-number tandem-repeat analysis (MLVA), and macrolide susceptibility genotyping.
Results. Of 216 M pneumoniae polymerase chain reaction-positive specimens, 40 (18.5%) were obtained from adults and 176 (81.5%) from children. P1 type distribution differed between adults (64% type 1 and 36% type 2) and children (84% type 1, 13% type 2, and 3% variant) (P < .05) and among sites (P < .01). Significant differences in the proportions of MLVA types 4/5/7/2 and 3/5/6/2 were also observed by age group (P < .01) and site (P < .01). A macrolide-resistant genotype was identified in 7 (3.5%) specimens, 5 of which were from patients who had recently received macrolide therapy. No significant differences in clinical characteristics were identified among patients with various strain types or between macrolide-resistant and -sensitive M pneumoniae infections.
Conclusions. The P1 type 1 genotype and MLVA type 4/5/7/2 predominated, but there were differences between children and adults and among sites. Macrolide resistance was rare. Differences in strain types did not appear to be associated with differences in clinical outcomes. Whole genome sequencing of M pneumoniae may help identify better ways to characterize strains.
Mycoplasma pneumoniae is a common cause of respiratory infections, including community-acquired pneumonia (CAP). However, the disease burden is difficult to estimate due to limitations of diagnostic assays and lack of systematic surveillance [1, 2]. Real-time polymerase chain reaction (PCR) for M pneumoniae detection is preferred due to the improved sensitivity and specificity compared with culture and serology, yet routine PCR testing in the clinical setting remains uncommon [2].
Molecular characterization of circulating M pneumoniae strains in the United States is particularly limited. Classification as type 1, type 2, or variant genotypes based on sequence variation in the gene encoding the immunogenic P1 surface protein has been the standard method for differentiating M pneumoniae [1–3]. More recently, the development of a multilocus variable-number tandem-repeat analysis (MLVA) typing scheme has allowed more precise categorization of strains based on the variable copy number of tandemly repeated sequences at multiple stable genetic loci [4, 5]. However, the clinical utility of P1 and MLVA typing is uncertain because no genotype has been associated with increased virulence or epidemic potential.
Advanced molecular methods are also useful for identifying genetic mutations conferring macrolide resistance in M pneumoniae, a growing global public health concern [6, 7]. Some studies suggest that individuals infected with macrolide-resistant M pneumoniae experience a longer febrile period, more persistent cough, and extended antibiotic therapy compared with persons infected with macrolide-sensitive strains [8–13], although the impact of macrolide resistance on patient outcome remains uncertain. In Asia, over 90% of isolates are resistant to macrolides [14, 15]. Resistance has also emerged in the United States over the last 15 years [7, 16, 17], but the prevalence is unknown.
We characterized M pneumoniae detections from a cohort of adults and children hospitalized with radiographically confirmed CAP prospectively enrolled in the Centers for Disease Control and Prevention (CDC) Etiology of Pneumonia in the Community (EPIC) study.
METHODS
Study Population, Case Definitions, and Clinical Specimens
Children (<18 years old) and adults were enrolled in the EPIC study from January 2010 to June 2012 at 8 hospitals in Chicago, Illinois, Memphis, Tennessee, Nashville, Tennessee, and Salt Lake City, Utah [18]. Adults were enrolled in Chicago and Nashville; children were enrolled in Nashville, Memphis, and Salt Lake City. Informed consent was obtained before enrollment. The study protocol was approved by the institutional review boards at each institution and the CDC. Individuals admitted to a study hospital with evidence of acute respiratory infection and radiographic confirmation of pneumonia were included: patients who were recently hospitalized or severely immunocompromised were excluded [18]. Combined nasopharyngeal/oropharyngeal (NP/OP) swabs were obtained from all patients for molecular detection of respiratory viruses and atypical bacteria, including M pneumoniae [18]. Testing for M pneumoniae was performed using an individual real-time PCR assay designed to detect the community-acquired respiratory distress syndrome (CARDS) toxin gene and validated by the CDC Pneumonia Response and Surveillance Laboratory (PRSL) [19]. For this analysis, cases were defined as enrolled patients meeting the final CAP case definition [18] with an M pneumoniae-positive PCR result (crossing threshold [Ct] value <40) from a NP/OP specimen collected within 72 hours of admission. Additional respiratory and blood specimens were also collected from adults and children (and urine for adults only) for bacterial and viral testing as previously described (Supplementary Material) [18].
Specimen Processing
After initial testing at study sites, NP/OP specimens were stored at ≤−70°C and shipped to the CDC for long-term storage. All M pneumoniae PCR-positive specimens were transferred to PRSL for additional molecular testing. Total nucleic acid (TNA) was extracted using the MagNA Pure Compact System with Total Nucleic Acid Isolation Kit I (Roche Applied Science) according to manufacturer's instructions. All M pneumoniae-positive specimens were tested at CDC using a validated multiplex PCR assay for detection of M pneumoniae (CARDS toxin gene), Chlamydophila pneumoniae, Legionella spp, and human RNaseP (internal control) to confirm the initial positive M pneumoniae PCR result obtained at the study site [20]; no Ct value cutoff was used for confirmatory testing. Various testing methodologies, including culture, PCR, antigen detection, and serology, were performed for detection of other bacteria and viruses in other specimen types collected from these patients per the EPIC study diagnostic algorithm as previously described (Supplementary Material).
Culture
Culture was attempted on all M pneumoniae PCR-positive specimens using SP4 medium (Remel) as previously described to obtain isolates for testing [21]. Nucleic acid was extracted from liquid culture, and recovery of M pneumoniae was confirmed by individual singleplex real-time PCR assay [19].
P1 Subtyping
Genotyping of the P1 adhesin gene was attempted for all isolates recovered from culture using real-time PCR with high-resolution melt (HRM) as previously described [22]. Isolates were classified as type 1, type 2, or variant genotypes based on comparison of the HRM profile to reference strains M129 (type 1) and FH (type 2) included in each run [3, 22]. Because P1 typing of primary specimen extracts is unreliable, this assay was performed only on culture isolates using a normalized concentration of nucleic acid.
Multilocus Variable-Number Tandem-Repeat Analysis
Multilocus variable-number tandem-repeat analysis was performed on nucleic acid extracts from all primary specimens and isolates as previously described [4, 5]. Multilocus variable-number tandem-repeat analysis types were reported using the modified 4 variable-number tandem-repeat (VNTR) loci method; the Mpn1 locus was excluded due to documented instability [4, 23].
Macrolide Susceptibility
Macrolide susceptibility testing was performed on all M pneumoniae PCR-positive specimens and corresponding culture isolates by genotyping of the 23S rRNA gene using a real-time PCR assay with HRM analysis as previously described [7, 17]. This method allows detection of an A to G transition at position 2063 or 2064 within the 23S rRNA gene, the 2 mutations most commonly associated with macrolide resistance in M pneumoniae [7, 10, 24]. High-resolution melt profiles were compared with sensitive and resistant reference strains included in each run and previously confirmed by sequencing analysis and minimum inhibitory concentration determination [7]. Sequencing analysis was performed on all macrolide-resistant isolates to identify the specific single-base mutation (A2063G or A2064G) in the 23S rRNA gene [7]. Prior exposure to macrolide antibiotics, defined as azithromycin or clarithromycin received 1–13 days before enrollment, was determined by patient interviews and medical chart abstraction.
Analysis
The proportions of macrolide-resistant M pneumoniae, P1 subtypes, and MLVA types were compared between age groups (adults vs children), enrollment city, and select clinical characteristics using χ2 or Fisher's exact test as appropriate. All analyses were conducted using SAS version 9.3 (Cary, NC); P < .05 was considered significant.
RESULTS
Characteristics of Patients With Mycoplasma pneumoniae Community-Acquired Pneumonia
Among 225 M pneumoniae PCR-positive specimens received from the study hospitals, 9 (4%) were negative for M pneumoniae upon repeat real-time PCR testing at the CDC and were excluded (Supplementary Figure 1). Mycoplasma pneumoniae isolates were recovered by culture from 175 (81%) of the 216 confirmed PCR-positive specimens (Supplementary Figure 1). Among 216 patients having an M pneumoniae PCR-positive NP/OP specimen, 24.5% had another bacterial or viral pathogen detected, including 7.5% of adults and 28.5% of children (Supplementary Table 1). Chlamydia pneumoniae and Legionella spp were not detected in any specimens upon confirmatory PCR testing at the CDC.
Of the 216 M pneumoniae PCR-positive specimens included in this analysis, 40 (18.5%) were obtained from adults and 176 (81.5%) were from children (Table 1). Among these patients, 4 (10.0%) of 40 adults and 19 (10.8%) of 176 children required intensive care unit (ICU) admission; there were no deaths. No adults and 3 (1.7%) of 176 children required invasive mechanical ventilation. Median length of stay was 2.5 days for adults (interquartile range [IQR], 1.5–4) and 2.0 days (IQR, 2.5–4) for children (data not shown).
Table 1.
Characteristics of Patients With Mycoplasma pneumoniae-Positive NP/OP Specimensa
| Characteristic | Total (n = 216), n (%) | Adults (n = 40), n (%) | Children (n = 176), n (%) |
|---|---|---|---|
| Site | |||
| Chicago | 30 (13.9) | 30 (75.0) | N/Aa |
| Memphis | 59 (27.3) | N/A | 59 (33.5) |
| Nashville | 47 (21.8) | 10 (25.0) | 37 (21.0) |
| Salt Lake City | 80 (37.0) | N/A | 80 (45.5) |
| Age Group | |||
| 0–23 months | 17 (7.9) | – | 17 (9.7) |
| 2–4 years | 30 (13.9) | – | 30 (17.0) |
| 5–9 years | 67 (31.0) | – | 67 (38.1) |
| 10–17 years | 62 (28.7) | – | 62 (35.2) |
| 18–49 years | 25 (11.6) | 25 (62.5) | – |
| 50–64 years | 7 (3.2) | 7 (17.5) | – |
| 65–79 years | 6 (2.8) | 6 (15.0) | – |
| ≥80 years | 2 (0.9) | 2 (5.0) | – |
| Gender | |||
| Male | 124 (57.4) | 19 (47.5) | 105 (59.7) |
| Female | 92 (42.6) | 21 (52.5) | 71 (40.3) |
| Race/Ethnicity | |||
| Non-Hispanic White | 131 (60.6) | 22 (55.0) | 109 (61.9) |
| Non-Hispanic Black | 41 (19) | 10 (25.0) | 31 (17.6) |
| Hispanic | 36 (16.7) | 8 (20) | 28 (15.9) |
| Other | 8 (3.7) | 0 (0) | 8 (4.5) |
Abbreviations: N/A, not applicable; NP/OP, nasopharyngeal/oropharyngeal.
a Adults were enrolled at Chicago and Nashville. Children were enrolled at Nashville, Memphis, and Salt Lake City.
P1 Genotyping and Multilocus Variable-Number Tandem-Repeat Analysis
The 2 main P1 genotypes of M pneumoniae, types 1 and 2, accounted for 81.1% and 16.6% of total cultured isolates (n = 175), respectively (Table 2). Type 1 accounted for the majority of detections across all ages, including 64.3% of detections in adults and 84.4% in children, whereas type 2 accounted for 35.7% in adults and 12.9% in children (P < .01). Four variant strains (2.3%) were also identified, all in pediatric specimens.
Table 2.
Molecular Characteristics of Mycoplasma pneumoniae by Age Group
| Characteristic | Total (n = 216), n (%) | Adults (n = 40), n (%) | Children (n = 176), n (%) | P Valuea |
|---|---|---|---|---|
| Macrolide profileb | n = 202 | n = 33 | n = 169 | 1.0 |
| Sensitive | 195 (96.5) | 32 (97.0) | 163 (96.4) | |
| Resistant | 7 (3.5) | 1 (3.0) | 6 (3.6) | |
| P1 genotypec | n = 175 | n = 28 | n = 147 | .02 |
| Type 1 | 142 (81.1) | 18 (64.3) | 124 (84.4) | |
| Type 2 | 29 (16.6) | 10 (35.7) | 19 (12.9) | |
| Variant | 4 (2.3) | 0 (0) | 4 (2.7) | |
| MLVA typed | n = 208 | n = 37 | n = 171 | <.01 |
| 4/5/7/2 | 149 (71.6) | 18 (48.6) | 131 (76.6) | |
| 3/5/6/2 | 33 (15.9) | 13 (35.1) | 20 (11.7) | |
| Other | 26 (12.5) | 6 (16.2) | 20 (11.7) | |
| 3/4/6/2 | 1 (0.5) | 1 (2.7) | 0 (0) | |
| 3/6/6/2 | 9 (4.3) | 2 (5.4) | 7 (4.1) | |
| 4/0/7/2 | 2 (1.0) | 0 (0) | 2 (1.2) | |
| 4/5/6/2 | 2 (1.0) | 1 (2.7) | 1 (0.6) | |
| 4/5/7/0 | 5 (2.4) | 1 (2.7) | 4 (2.3) | |
| 4/6/7/2 | 6 (2.9) | 1 (2.7) | 5 (2.9) | |
| 5/5/7/0 | 1 (0.5) | 0 (0) | 1 (0.6) |
Abbreviations: MLVA, multilocus variable-number tandem-repeat analysis.
a The χ2 test or Fisher's exact test as appropriate comparing children with adults.
b Macrolide profile could not be determined for 14 (6.5%) of 216 specimens, including 7 (17.5%) of adults and 7 (4%) of children, due to poor amplification of target sequence from primary specimen and/or lack of isolate recovery.
c P1 genotype was determined for culture isolates only (n = 175).
d MLVA type could not be determined for 8 (3.7%) of 216 specimens, including 3 (7.5%) of adults and 5 (2.8%) of children, due to poor amplification of target sequence from primary specimen and/or lack of isolate recovery. Other types shown were grouped for statistical comparison with the predominant types 4/5/7/2 and 3/5/6/2.
Multilocus variable-number tandem-repeat analysis type could not be determined for 8 (3.7%) of 216 specimens due to poor amplification of the target sequence from the primary specimen and lack of isolate recovery (Supplementary Figure 1). Nine distinct MLVA types were identified among the remaining 208 M pneumoniae PCR-positive specimens (Tables 2 and 3). The majority (71.6%) of detections were type 4/5/7/2, including 48.6% of adults and 76.6% of children. Type 3/5/6/2 was the second most commonly identified MLVA type in both adults (35.1%) and children (11.7%). There were no significant differences in ICU admission, invasive mechanical ventilation, length of stay, or the proportion of other bacterial or viral pathogen detections among P1 genotypes or MLVA types (Supplementary Tables 1 and 2).
Table 3.
Molecular Characteristics of Mycoplasma pneumoniae by Site
| Characteristic | Chicago (n = 30), n (%) |
Memphis (n = 59), n (%) |
Nashville (n = 47), n (%) |
Salt Lake City (n = 80), n (%) |
P Valuea |
|---|---|---|---|---|---|
| Macrolide profileb | n = 23 | n = 59 | n = 46 | n = 74 | .3 |
| Sensitive | 23 (100) | 56 (94.9) | 43 (93.5) | 73 (98.6) | |
| Resistant | 0 (0) | 3 (5.1) | 3 (6.5) | 1 (1.4) | |
| P1 genotypec | n = 20 | n = 56 | n = 33 | n = 66 | <.01 |
| Type 1 | 12 (60.0) | 42 (75.0) | 26 (78.8) | 62 (93.9) | |
| Type 2 | 8 (40.0) | 11 (19.6) | 6 (18.2) | 4 (6.1) | |
| Variant | 0 (0) | 3 (5.6) | 1 (3.0) | 0 (0) | |
| MLVA typed | n = 28 | n = 57 | n = 44 | n = 79 | <.01 |
| 4/5/7/2 | 13 (46.4) | 43 (75.4) | 30 (68.2) | 63 (79.7) | |
| 3/5/6/2 | 10 (35.7) | 10 (17.5) | 9 (20.5) | 4 (5.1) | |
| Other | 5 (17.9) | 4 (7.0) | 5 (11.4) | 12 (17.7) | |
| 3/4/6/2 | 1 (3.6) | 0 (0) | 0 (0) | 0 (0) | |
| 3/6/6/2 | 2 (7.1) | 4 (7.0) | 3 (6.8) | 0 (0) | |
| 4/0/7/2 | 0 (0) | 0 (0) | 0 (0) | 2 (2.5) | |
| 4/5/6/2 | 0 (0) | 0 (0) | 2 (4.5) | 0 (0) | |
| 4/5/7/0 | 1 (3.6) | 0 (0) | 0 (0) | 4 (5.1) | |
| 4/6/7/2 | 1 (3.6) | 0 (0) | 0 (0) | 5 (6.3) | |
| 5/5/7/0 | 0 (0) | 0 (0) | 0 (0) | 1 (1.3) |
Abbreviations: MLVA, multilocus variable-number tandem-repeat analysis.
a The χ2 test or Fisher's exact test comparing all 4 cities.
b Macrolide profile could not be determined for 14 (6.5%) of 216 specimens due to poor amplification of target sequence from primary specimen and/or lack of isolate recovery.
c P1 genotype was determined for isolates only (n = 175).
d MLVA type could not be determined for 8 (3.7%) of 216 specimens due to poor amplification of target sequence from primary specimen and/or lack of isolate recovery. Other types shown were grouped for statistical comparison with the predominant types 4/5/7/2 and 3/5/6/2.
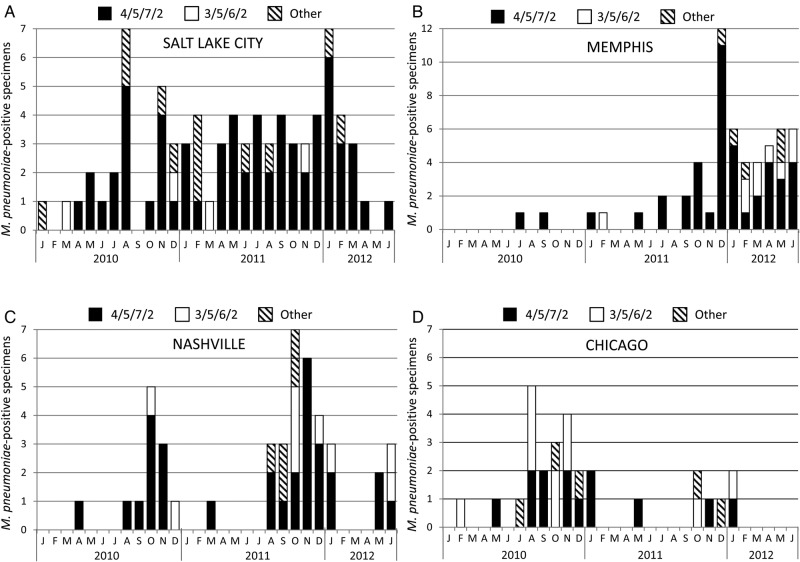
The distribution of P1 genotypes and MLVA types differed significantly between adults and children (P < .05; Table 2) and among sites (P < .01, Table 3). Multilocus variable-number tandem-repeat analysis type 4/5/7/2 predominated at each site across the entire study period except in Chicago (adults only) where type 3/5/6/2 was also common (Figure 1 and Table 3). The proportions of MLVA types 4/5/7/2, 3/5/6/2, and other types were similar in adults from Nashville and Chicago (Table 3 and Supplementary Figure 2); P1 type distribution was also similar among adults at these 2 sites (Chicago: 60% type 1 and 40% type 2; Nashville: 75% type 1 and 25% type 2). Likewise, the proportions of P1 types (80% type 1, 16% type 2, 4% variant) and MLVA types (Supplementary Figure 2) among children enrolled in Nashville were similar to those observed at the other pediatric enrollment sites (Table 3). In Nashville, the only site enrolling both adults and children, there were no significant differences in the proportions of MLVA types or P1 types between age groups. Two MLVA types, 4/0/7/2 (n = 2) and 5/5/7/0 (n = 1), were identified only in children, whereas type 3/4/6/2 was identified in a single adult specimen (Table 2). Three novel types were identified, including 4/5/7/0 and 5/5/7/0, which lacked the Mpn16 locus, and 4/0/7/2, which lacked the Mpn14 locus. Multilocus variable-number tandem-repeat analysis results were confirmed by repeating the test with a normalized concentration of TNA from the culture isolate to ensure that the lack of amplification of an individual locus was not due to limited nucleic acid in the primary specimen.
Figure 1.
Number of Mycoplasma pneumoniae detections of the 2 predominant multilocus variable-number tandem-repeat analysis (MLVA) types, 4/5/7/2 (black bars) and 3/5/6/2 (white bars), and other types (hashed bars) from children in Salt Lake City (A) and Memphis (B), children and adults in Nashville (C), and adults in Chicago (D) over the study period. Other category includes the following: 3/6/6/2, 3/4/6/2, 4/0/7/2, 4/5/6/2, 4/5/7/0, 4/6/7/2, and 5/5/7/0. Note that the y-axis scale for Memphis (B) differs from the other graphs. Detections of each MLVA type in adults and children in Nashville are shown separately in Supplementary Figure 2.
Macrolide Susceptibility
The macrolide susceptibility genotype could not be determined for 14 (6.5%) of the 216 M pneumoniae PCR-positive specimens, including 7 (17.5%) adults and 7 (4%) children, due to poor amplification of the target sequence from the primary specimen and inability to culture the organism (Supplementary Figure 1). Of the remaining 202 specimens, 195 (96.5%) were sensitive to macrolides and 7 (3.5%) were resistant (Table 2). The proportions of predominant MLVA types were similar among resistant isolates (86% type 4/5/7/2 and 14% type 3/6/6/2) and all specimens (72% type 4/5/7/2 and 16% type 3/6/6/2) (Tables 2 and 4). Sequencing analysis revealed the presence of the A2063G mutation in 6 isolates and A2064G in 1 isolate.
Table 4.
Characteristics of Macrolide-Resistant Mycoplasma pneumoniae (n = 7)
| Site | n (%) |
|---|---|
| Chicago | 0 (0) |
| Memphis | 3 (43) |
| Nashville | 3 (43) |
| Salt Lake City | 1 (14) |
| Specimen collection year | |
| 2010 | 1 (14.3) |
| 2011 | 5 (71.4) |
| 2012 | 1 (14.3) |
| Patient age | |
| 0–23 months | 1 (14) |
| 2–4 years | 0 (0) |
| 5–9 years | 2 (29) |
| 10–17 years | 3 (43) |
| 18–49 years | 1 (14) |
| ≥50 years | 0 (0) |
| Timing of macrolide relative to specimen collection | |
| Before | 5 (71%) |
| Aftera | 2 (29%) |
| 23S rRNA genotype | |
| A2063G | 6 (86) |
| A2064G | 1 (14) |
| P1 genotype | |
| Type 1 | 6 (86) |
| Type 2 | 0 (0) |
| Variant | 1 (14) |
| MLVA type | |
| 3/6/6/2 | 1 (14) |
| 4/5/7/2 | 6 (86) |
Abbreviations: MLVA, multilocus variable-number tandem-repeat analysis.
a Includes macrolide received on same day as specimen collection (n = 2).
The proportion of resistant M pneumoniae did not differ significantly between adults (3.0%) and children (3.6%). Characteristics of the 7 macrolide-resistant isolates are shown in Table 4. At least 1 resistant isolate was identified at all 3 pediatric sites (Table 3). Five (71%) of 7 resistant isolates were recovered from patients who received a macrolide for the current illness before specimen collection compared with 44 (23%) of 195 susceptible specimens (P < .01). Among patients with resistant M pneumoniae isolates, none required ICU admission or invasive mechanical ventilation, and median length of stay was 3.0 days (IQR, 1–6) compared with 2.0 days (IQR, 2–4) for patients with macrolide-sensitive M pneumoniae (Supplementary Table 2).
DISCUSSION
This study is a comprehensive molecular analysis of M pneumoniae-associated CAP in both children and adults at multiple sites within the United States. P1 type 1 and MLVA type 4/5/7/2 were most common in our study, which is consistent with recent reports from the United States and other regions of the world [17, 23]. A single MLVA type, 4/5/7/2, predominated in children, whereas 2 MLVA types, 4/5/7/2 and 3/5/6/2, were common in adults. In addition, in this study of hospitalized US adults and children using a strict definition of CAP, M pneumoniae macrolide genotypic resistance was low. No significant differences in clinical characteristics were identified among patients with varying strain types or between macrolide-resistant and sensitive M pneumoniae infections.
This study occurred simultaneously with a reported increase in M pneumoniae transmission in Europe and Asia during 2010–2012 [23, 25]. Four M pneumoniae outbreaks in the United States and several sporadic cases or clusters were investigated by CDC in 2013, which was an increase from previous years [17]. Increased detection of M pneumoniae in the United States could be partially explained by broader implementation of molecular diagnostics in clinical settings [26, 27]. However, reports of increased detections on multiple continents suggest that 2010–2013 represented a period of increased M pneumoniae transmission, which occur regularly every 3 to 7 years [1, 28]. Although this periodicity of M pneumoniae has been attributed to continual re-emergence of the less prevalent P1 type due to waning immunity within the population over time [28], numerous recent reports have described co-circulation of multiple P1 and MLVA types within a population or during an outbreak [5, 17, 29, 30]. Co-circulation of the 2 main P1 types was also observed in each city in the current study. Implementation of a systematic surveillance program incorporating molecular characterization may be useful for determining whether changes in M pneumoniae disease burden correlate with changes in distribution of strain types within the population.
Using a modified typing method based on 4 VNTR loci, 15 unique MLVA types have been described to date [23]. The most prevalent MLVA types among EPIC specimens were 4/5/7/2 and 3/5/6/2, which is consistent with recent analyses of circulating MLVA types within the United States and internationally [4, 5, 23, 25, 31–33]. Significant differences were identified in the proportions of MLVA types and P1 types between adults and children as well as among sites: MLVA type 4/5/7/2 was predominant in all age groups and at all sites, but 3/5/6/2 was more common in adults than children. However, in Nashville, the only site enrolling both adults and children, there were no significant differences in the proportions of MLVA types or P1 types between age groups. Thus, the observed differences in distribution of strain types may be attributed to geography, patient age, or a combination of factors. Although co-detections of other bacterial or viral or pathogens were observed, mostly in children, no differences in presence of co-detections were observed based on P1 genotype or MLVA type.
A correlation between the 2 main typing methods has previously been demonstrated such that P1 type can be predicted by MLVA profile; type 4/5/7/2 strains are P1 type 1, whereas type 3/5/6/2 strains reliably have the P1 type 2 genotype [4, 17, 34]. This correlation was also observed among M pneumoniae PCR-positive specimens in the current study (data not shown). Therefore, MLVA typing may have greater utility because it affords the ability to further distinguish strains while still predicting P1 type. Several previously unreported MLVA types lacking either the Mpn14 or Mpn16 locus were discovered in this analysis. Further sequence analysis of these strains is needed to identify whether the entire locus is deleted or otherwise modified and what implications this may have for pathogenesis, transmission, or disease outcomes. Furthermore, no significant differences in clinical characteristics were observed among this cohort of patients based on strain type. Our results indicate that current typing methods are insufficient to meaningfully differentiate M pneumoniae because the majority of strains can be categorized into only a few main types, none of which have been identified as clinically informative. Whole genome sequencing of M pneumoniae may result in improved methods for strain characterization.
Macrolide resistance was identified in only 3.5% of M pneumoniae PCR-positive specimens in this study. This prevalence is consistent with the previously reported prevalence of ≤10% seen during surveillance studies and outbreak investigations in the United States, although few reports are available for comparison [6, 16, 35]. Previous reports were limited in sample size and geographic coverage, were based on clinical testing practices, and included both hospitalized patients and those with mild disease [16, 17]. Our prospective, population-based analysis included 4 geographically distinct cities enrolling adults, children, or both age groups, and thus it represents the most comprehensive examination of macrolide resistance among hospitalized patients with M pneumoniae CAP in the United States to date.
The proportions of macrolide-resistant M pneumoniae reported in the United States are generally consistent with those observed from routine surveillance in northern Europe, varying between 3% and 10% [25, 29, 36]. In contrast, macrolide resistance dramatically increased in Japan and China during the past decade to over 90% in some reports, underscoring the potential for rapid emergence of antimicrobial resistance within M pneumoniae [11, 14, 15, 37]. Efforts to reduce unnecessary antibiotic prescribing and inappropriate antibiotic selection for respiratory infections could help prevent emergence of widespread macrolide resistance in North America [38]. Although only 7 resistant isolates were identified, macrolide resistance was associated with recent receipt of a macrolide before study enrollment, supporting the theory that a resistant subpopulation may develop or expand during the course of macrolide therapy within an individual patient [13, 39, 40]. The proportions of predominant MLVA types were similar between resistant and susceptible isolates, suggesting that resistance is not more likely to develop in a specific MLVA type. Further laboratory and epidemiological studies are needed to understand the effect of macrolide exposure and potential mechanisms for selection or development of resistant M pneumoniae in response to macrolide treatment.
There are several limitations to this analysis. Inadequate amplification of target regions resulting in inconclusive MLVA and macrolide genotyping was most likely due to low quantity of pathogen-specific nucleic acid in the primary specimen as suggested by high Ct values upon initial specimen testing (Supplementary Figure 1). Although this was a multicenter study, the microbiological characteristics of M pneumoniae, including the prevalence of macrolide resistance, may differ in other regions of the United States not represented in our study. Furthermore, enrollment of adults and children occurred only at 1 of the 4 sites, precluding definitive attribution of significant differences in strain type distribution to either age or geography. Macrolide susceptibility testing was performed on M pneumoniae-positive specimens collected within 72 hours of admission. Thus, resistance mutations induced as a result of subsequent treatment were not detected.
This study provides a deeper understanding of M pneumoniae biology, molecular epidemiology, and macrolide resistance among patients hospitalized with CAP. Further investigation is also warranted to understand the biological and epidemiological reasons that may explain the differences in distribution of M pneumoniae types between adult and pediatric populations. Investigation of M pneumoniae using next-generation sequencing may (1) provide further characterization and insight into the evolution of M pneumoniae within the human host population and (2) afford the opportunity to improve upon current typing methods. Further studies are warranted to understand the full spectrum of M pneumoniae illness, monitor the emergence of antibiotic resistance, and define specific microbial determinants of pathogenesis.
Supplementary Material
Supplementary material is available online at Open Forum Infectious Diseases (http://OpenForumInfectiousDiseases.oxfordjournals.org/).
Acknowledgments
We thank the patients who graciously consented to participate in this study and all members of the Etiology of Pneumonia in the Community Study Team for their contributions.
Disclaimers. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Financial support. This work was supported by the Influenza Division in the National Center for Immunization and Respiratory Diseases at the Centers for Disease Control and Prevention (CDC) through cooperative agreements with each study site and was based on a competitive research funding opportunity.
Potential conflicts of interest. J. D. C. received grants from the CDC during the conduct of the study and has a patent licensed to Vanderbilt University (US patent 8 293 498 B2, October 23, 2012) and a patent pending (Provisional application for US letters patent, VBLT:171USP1). D. J. W. received grants from the CDC during the conduct of the study. S. R. A. received grants from the CDC during the conduct of the study and received grants from GlaxoSmithKline outside the submitted work. K. A. received grants from the CDC during the conduct of the study and collaborated with BioFire Diagnostics, Inc. W. H. S. received grants from the CDC during the conduct of the study; received research supplies from CareFusion; received grants from BioMerieux, Affinium Pharmaceuticals, Astute Medical, Crucell Holland BV, BRAHMS GmbH, Pfizer, Rapid Pathogen Screening, Venaxis, Cempra Pharmaceuticals, BioAegis Inc, and Sphingotec GmbH; received personal fees from BioFire Diagnostics and Venaxis, Inc. outside the submitted work; and has a patent pending (13/632,874). E. J. A. received grants from MedImmune and nonfinancial support from Roche outside the submitted work. J. A. M. received grants from the CDC during the conduct of the study. A. T. P. received grants from the CDC during the conduct of the study; and travel support and consultant fee from BioFire Diagnostics outside the submitted work. R. G. W. received grants from the CDC during the conduct of the study and received grants and personal fees from BioMerieux Inc and personal fees from Cempra Pharmaceuticals. K. M. E. received grants from the CDC during the conduct of the study and received grants from Novartis and funding for service on a Data and Safety Monitoring Board from Novartis outside the submitted work.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Atkinson TP, Balish MF, Waites KB. Epidemiology, clinical manifestations, pathogenesis and laboratory detection of Mycoplasma pneumoniae infections. FEMS Microbiol Rev 2008; 32:956–73. [DOI] [PubMed] [Google Scholar]
- 2.Winchell JM. Mycoplasma pneumoniae - A National Public Health Perspective. Curr Pediatr Rev 2013; 9:324–33. [Google Scholar]
- 3.Schwartz SB, Mitchell SL, Thurman KA et al. Identification of P1 variants of Mycoplasma pneumoniae by use of high-resolution melt analysis. J Clin Microbiol 2009; 47:4117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benitez AJ, Diaz MH, Wolff BJ et al. Multilocus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical isolates from 1962 to the present: a retrospective study. J Clin Microbiol 2012; 50:3620–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Degrange S, Cazanave C, Charron A et al. Development of multiple-locus variable-number tandem-repeat analysis for the molecular typing of Mycoplasma pneumoniae. J Clin Microbiol 2009; 47:914–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Atkinson TP, Hagood J et al. Emerging macrolide resistance in Mycoplasma pneumoniae in children: detection and characterization of resistant isolates. Pediatr Infect Dis J 2009; 28:693–6. [DOI] [PubMed] [Google Scholar]
- 7.Wolff BJ, Thacker WL, Schwartz SB, Winchell JM. Detection of macrolide resistance in Mycoplasma pneumoniae by real-time PCR and high-resolution melt analysis. Antimicrob Agents Chemother 2008; 52:3542–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao B, Zhao CJ, Yin YD et al. High prevalence of macrolide resistance in Mycoplasma pneumoniae isolates from adult and adolescent patients with respiratory tract infection in China. Clin Infect Dis 2010; 51:189–94. [DOI] [PubMed] [Google Scholar]
- 9.Matsubara K, Morozumi M, Okada T et al. A comparative clinical study of macrolide-sensitive and macrolide-resistant Mycoplasma pneumoniae infections in pediatric patients. J Infect Chemother 2009; 15:380–3. [DOI] [PubMed] [Google Scholar]
- 10.Matsuoka M, Narita M, Okazaki N et al. Characterization and molecular analysis of macrolide-resistant Mycoplasma pneumoniae clinical isolates obtained in Japan. Antimicrob Agents Chemother 2004; 48:4624–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morozumi M, Iwata S, Hasegawa K et al. Increased macrolide resistance of Mycoplasma pneumoniae in pediatric patients with community-acquired pneumonia. Antimicrob Agents Chemother 2008; 52:348–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki S, Yamazaki T, Narita M et al. Clinical evaluation of macrolide-resistant Mycoplasma pneumoniae. Antimicrob Agents Chemother 2006; 50:709–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardinale F, Chironna M, Chinellato I et al. Clinical relevance of Mycoplasma pneumoniae macrolide resistance in children. J Clin Microbiol 2013; 51:723–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Ye X, Zhang H et al. Antimicrobial susceptibility of Mycoplasma pneumoniae isolates and molecular analysis of macrolide-resistant strains from Shanghai, China. Antimicrob Agents Chemother 2009; 53:2160–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xin D, Mi Z, Han X et al. Molecular mechanisms of macrolide resistance in clinical isolates of Mycoplasma pneumoniae from China. Antimicrob Agents Chemother 2009; 53:2158–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada M, Buller R, Bledsoe S, Storch GA. Rising rates of macrolide-resistant Mycoplasma pneumoniae in the central United States. Pediatr Infect Dis J 2012; 31:409–11. [DOI] [PubMed] [Google Scholar]
- 17.Diaz M, Benitez AJ, Winchell JM. Investigations of Mycoplasma pneumoniae infections in the United States: trends in molecular typing and macrolide resistance, 2006–2013. J Clin Microbiol 2015; 53:124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain S, Williams DJ, Arnold SR et al. Community-acquired pneumonia requiring hospitalization among U.S. children . N Engl J Med 2015; 372:835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winchell JM, Thurman KA, Mitchell SL et al. Evaluation of three real-time PCR assays for the detection of Mycoplasma pneumoniae in an outbreak investigation. J Clin Microbiol 2008; 46:3116–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thurman KA, Warner AK, Cowart KC et al. Detection of Mycoplasma pneumoniae, Chlamydia pneumoniae, and Legionella spp. in clinical specimens using a single-tube multiplex real-time PCR assay. Diagn Microbiol Infect Dis 2011; 70:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tully JG, Rose DL, Whitcomb RF, Wenzel RP. Enhanced isolation of Mycoplasma pneumoniae from throat washings with a newly modified culture medium. J Infect Dis 1979; 139:478–82. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz SB, Thurman KA, Mitchell SL et al. Genotyping of Mycoplasma pneumoniae isolates using real-time PCR and high-resolution melt analysis. Clin Microbiol Infect 2009; 15:756–62. [DOI] [PubMed] [Google Scholar]
- 23.Sun H, Xue G, Yan C et al. Multiple-locus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical specimens and proposal for amendment of MLVA nomenclature. PLoS One 2013; 8:e64607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bebear C, Pereyre S, Peuchant O. Mycoplasma pneumoniae: susceptibility and resistance to antibiotics. Future Microbiol 2011; 6:423–31. [DOI] [PubMed] [Google Scholar]
- 25.Steffens I, Hagmaier K, Wilson W, Wilson KE. Mycoplasmapneumoniae and Legionella pneumophila [Special edition]. 17th ed Sweden, Stockholm: Eurosurveillance, 2012. [Google Scholar]
- 26.Babady NE. The FilmArray(R) respiratory panel: an automated, broadly multiplexed molecular test for the rapid and accurate detection of respiratory pathogens. Expert Rev Mol Diagn 2013; 13:779–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratliff AE, Duffy LB, Waites KB. Comparison of the illumigene Mycoplasma DNA amplification assay and culture for detection of Mycoplasma pneumoniae. J Clin Microbiol 2014; 52:1060–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lind K, Benzon MW, Jensen S, Clyde WA Jr. A seroepidemiological study of Mycoplasma pneumoniae infections in Denmark over the 50-year period 1946 to1995. Eur J Epidemiol 1997; 13:581–6. [DOI] [PubMed] [Google Scholar]
- 29.Pereyre S, Charron A, Renaudin H et al. First report of macrolide-resistant strains and description of a novel nucleotide sequence variation in the P1 adhesin gene in Mycoplasma pneumoniae clinical strains isolated in France over 12 years. J Clin Microbiol 2007; 45:3534–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spuesens EB, Hoogenboezem T, Sluijter M et al. Macrolide resistance determination and molecular typing of Mycoplasma pneumoniae by pyrosequencing. J Microbiol Methods 2010; 82:214–22. [DOI] [PubMed] [Google Scholar]
- 31.Dumke R, Jacobs E. Culture-independent multi-locus variable-number tandem-repeat analysis (MLVA) of Mycoplasma pneumoniae. J Microbiol Methods 2011; 86:393–6. [DOI] [PubMed] [Google Scholar]
- 32.Pereyre S, Charron A, Hidalgo-Grass C et al. The spread of Mycoplasma pneumoniae is polyclonal in both an endemic setting in France and in an epidemic setting in Israel. PLoS One 2012; 7:e38585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao F, Liu G, Cao B et al. Multiple-locus variable-number tandem-repeat analysis of 201 Mycoplasma pneumoniae isolates from Beijing, China, from 2008 to 2011. J Clin Microbiol 2013; 51:636–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waller J, Diaz M, Petrone B et al. Detection and characterization of Mycoplasma pneumoniae during an outbreak of Respiratory illness at a university. J Clin Microbiol 2014; 52:849–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Critchley IA, Jones ME, Heinze PD et al. In vitro activity of levofloxacin against contemporary clinical isolates of Legionella pneumophila, Mycoplasma pneumoniae and Chlamydia pneumoniae from North America and Europe. Clin Microbiol Infect 2002; 8:214–21. [DOI] [PubMed] [Google Scholar]
- 36.Dumke R, von Baum H, Luck PC, Jacobs E. Occurrence of macrolide-resistant Mycoplasma pneumoniae strains in Germany. Clin Microbiol Infect 2010; 16:613–6. [DOI] [PubMed] [Google Scholar]
- 37.Zhao F, Liu G, Wu J et al. Surveillance of macrolide-resistant Mycoplasma pneumoniae in Beijing, China, from 2008 to 2012. Antimicrob Agents Chemother 2013; 57:1521–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCaig LF, Hicks LA, Roberts RM, Fairlie TA. Office-related antibiotic prescribing for persons aged </= 14 years--United States, 1993–1994 to 2007–2008. MMWR Morb Mortal Wkly Rep 2011; 60:1153–6. [PubMed] [Google Scholar]
- 39.Chironna M, Sallustio A, Esposito S et al. Emergence of macrolide-resistant strains during an outbreak of Mycoplasma pneumoniae infections in children. J Antimicrob Chemother 2011; 66:734–7. [DOI] [PubMed] [Google Scholar]
- 40.Dumke R, Stolz S, Jacobs E, Juretzek T. Molecular characterization of macrolide resistance of a Mycoplasma pneumoniae strain that developed during therapy of a patient with pneumonia. Int J Infect Dis 2014; 29C:197–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.